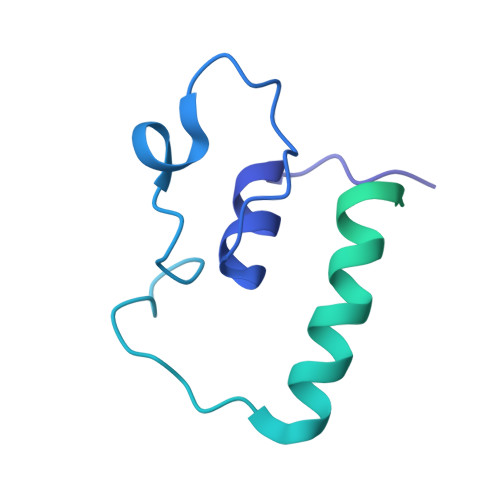
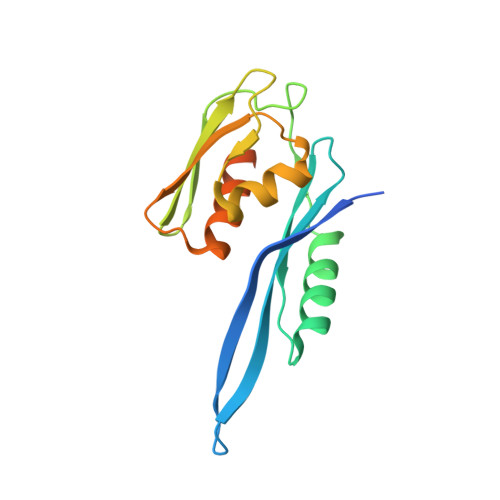
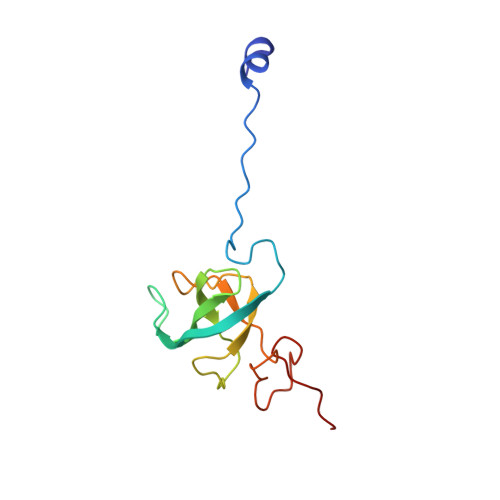
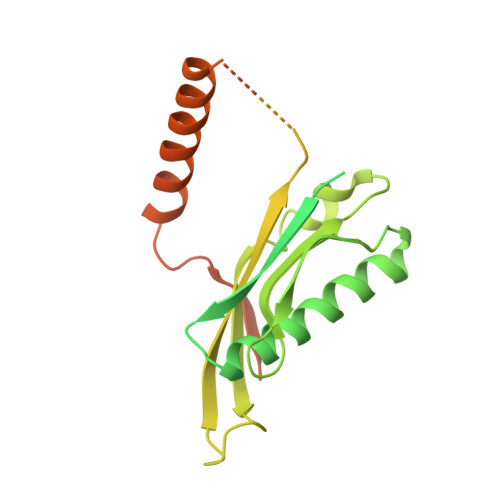
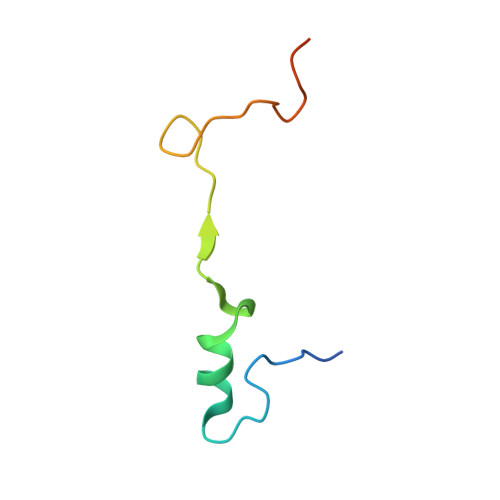
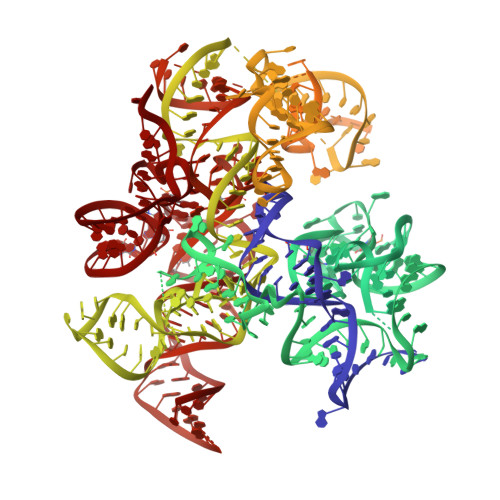
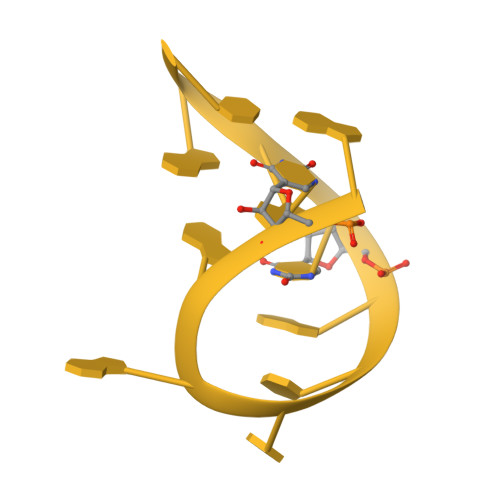
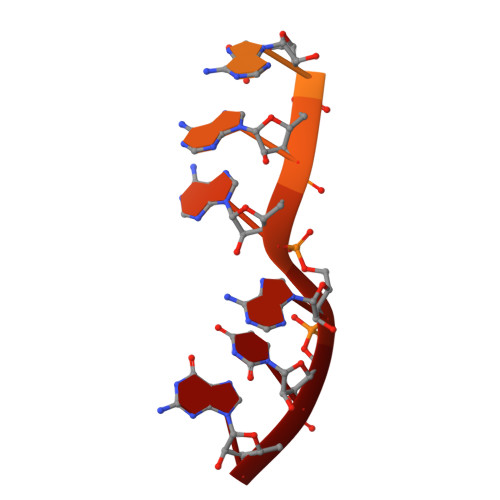
Structural basis of co-translational quality control by ArfA and RF2 bound to ribosome.
Zeng, F., Chen, Y., Remis, J., Shekhar, M., Phillips, J.C., Tajkhorshid, E., Jin, H.(2017) Nature 541: 554-557
- PubMed: 28077875
- DOI: https://doi.org/10.1038/nature21053
- Primary Citation of Related Structures:
5U4I, 5U4J - PubMed Abstract:
Quality control mechanisms intervene appropriately when defective translation events occur, in order to preserve the integrity of protein synthesis. Rescue of ribosomes translating on messenger RNAs that lack stop codons is one of the co-translational quality control pathways. In many bacteria, ArfA recognizes stalled ribosomes and recruits the release factor RF2, which catalyses the termination of protein synthesis. Although an induced-fit mechanism of nonstop mRNA surveillance mediated by ArfA and RF2 has been reported, the molecular interaction between ArfA and RF2 in the ribosome that is responsible for the mechanism is unknown. Here we report an electron cryo-microscopy structure of ArfA and RF2 in complex with the 70S ribosome bound to a nonstop mRNA. The structure, which is consistent with our kinetic and biochemical data, reveals the molecular interactions that enable ArfA to specifically recruit RF2, not RF1, into the ribosome and to enable RF2 to release the truncated protein product in this co-translational quality control pathway. The positively charged C-terminal domain of ArfA anchors in the mRNA entry channel of the ribosome. Furthermore, binding of ArfA and RF2 induces conformational changes in the ribosomal decoding centre that are similar to those seen in other protein-involved decoding processes. Specific interactions between residues in the N-terminal domain of ArfA and RF2 help RF2 to adopt a catalytically competent conformation for peptide release. Our findings provide a framework for understanding recognition of the translational state of the ribosome by new proteins, and expand our knowledge of the decoding potential of the ribosome.
- Department of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, USA.
Organizational Affiliation: