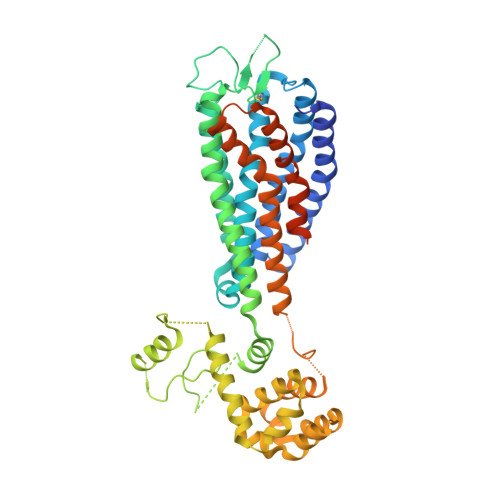
Structural basis for the cooperative allosteric activation of the free fatty acid receptor GPR40.
Lu, J., Byrne, N., Wang, J., Bricogne, G., Brown, F.K., Chobanian, H.R., Colletti, S.L., Di Salvo, J., Thomas-Fowlkes, B., Guo, Y., Hall, D.L., Hadix, J., Hastings, N.B., Hermes, J.D., Ho, T., Howard, A.D., Josien, H., Kornienko, M., Lumb, K.J., Miller, M.W., Patel, S.B., Pio, B., Plummer, C.W., Sherborne, B.S., Sheth, P., Souza, S., Tummala, S., Vonrhein, C., Webb, M., Allen, S.J., Johnston, J.M., Weinglass, A.B., Sharma, S., Soisson, S.M.(2017) Nat Struct Mol Biol 24: 570-577
- PubMed: 28581512
- DOI: https://doi.org/10.1038/nsmb.3417
- Primary Citation of Related Structures:
5TZR, 5TZY - PubMed Abstract:
Clinical studies indicate that partial agonists of the G-protein-coupled, free fatty acid receptor 1 GPR40 enhance glucose-dependent insulin secretion and represent a potential mechanism for the treatment of type 2 diabetes mellitus. Full allosteric agonists (AgoPAMs) of GPR40 bind to a site distinct from partial agonists and can provide additional efficacy. We report the 3.2-Å crystal structure of human GPR40 (hGPR40) in complex with both the partial agonist MK-8666 and an AgoPAM, which exposes a novel lipid-facing AgoPAM-binding pocket outside the transmembrane helical bundle. Comparison with an additional 2.2-Å structure of the hGPR40-MK-8666 binary complex reveals an induced-fit conformational coupling between the partial agonist and AgoPAM binding sites, involving rearrangements of the transmembrane helices 4 and 5 (TM4 and TM5) and transition of the intracellular loop 2 (ICL2) into a short helix. These conformational changes likely prime GPR40 to a more active-like state and explain the binding cooperativity between these ligands.
- Department of Structural Chemistry, Merck Research Laboratories, West Point, Pennsylvania, USA.
Organizational Affiliation: