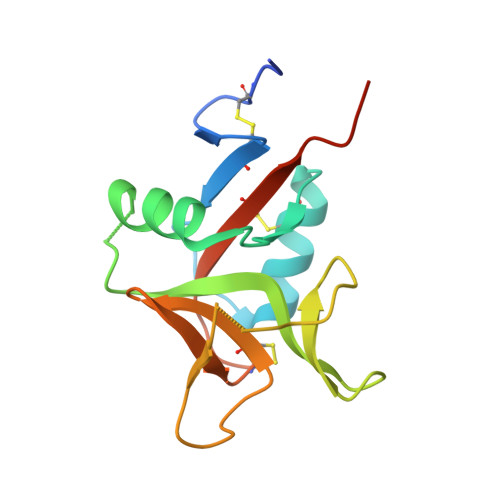
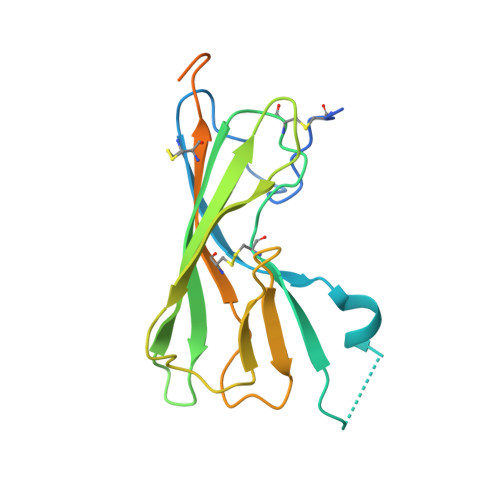
A Viral Immunoevasin Controls Innate Immunity by Targeting the Prototypical Natural Killer Cell Receptor Family.
Aguilar, O.A., Berry, R., Rahim, M.M., Reichel, J.J., Popovic, B., Tanaka, M., Fu, Z., Balaji, G.R., Lau, T.N., Tu, M.M., Kirkham, C.L., Mahmoud, A.B., Mesci, A., Krmpotic, A., Allan, D.S., Makrigiannis, A.P., Jonjic, S., Rossjohn, J., Carlyle, J.R.(2017) Cell 169: 58-71.e14
- PubMed: 28340350
- DOI: https://doi.org/10.1016/j.cell.2017.03.002
- Primary Citation of Related Structures:
5TZN - PubMed Abstract:
Natural killer (NK) cells play a key role in innate immunity by detecting alterations in self and non-self ligands via paired NK cell receptors (NKRs). Despite identification of numerous NKR-ligand interactions, physiological ligands for the prototypical NK1.1 orphan receptor remain elusive. Here, we identify a viral ligand for the inhibitory and activating NKR-P1 (NK1.1) receptors. This murine cytomegalovirus (MCMV)-encoded protein, m12, restrains NK cell effector function by directly engaging the inhibitory NKR-P1B receptor. However, m12 also interacts with the activating NKR-P1A/C receptors to counterbalance m12 decoy function. Structural analyses reveal that m12 sequesters a large NKR-P1 surface area via a "polar claw" mechanism. Polymorphisms in, and ablation of, the viral m12 protein and host NKR-P1B/C alleles impact NK cell responses in vivo. Thus, we identify the long-sought foreign ligand for this key immunoregulatory NKR family and reveal how it controls the evolutionary balance of immune recognition during host-pathogen interplay.
- Department of Immunology, University of Toronto, Toronto, ON M5S 1A8, Canada; Sunnybrook Research Institute, Toronto, ON M4N 3M5, Canada.
Organizational Affiliation: