Structural Basis of TPR-Mediated Oligomerization and Activation of Oncogenic Fusion Kinases.
Pal, K., Bandyopadhyay, A., Zhou, X.E., Xu, Q., Marciano, D.P., Brunzelle, J.S., Yerrum, S., Griffin, P.R., Vande Woude, G., Melcher, K., Xu, H.E.(2017) Structure 25: 867-877.e3
- PubMed: 28528776
- DOI: https://doi.org/10.1016/j.str.2017.04.015
- Primary Citation of Related Structures:
5TO5, 5TO6, 5TO7, 5TVB - PubMed Abstract:
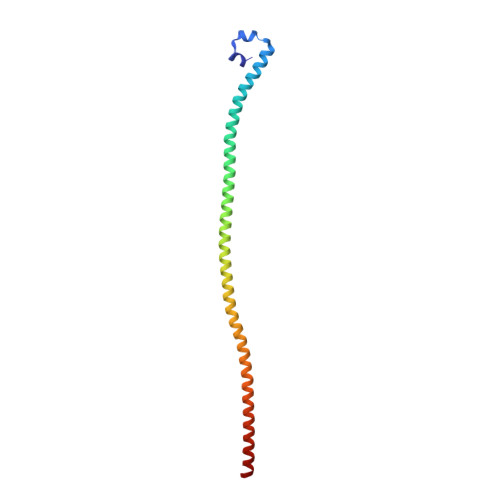
The nuclear pore complex subunit TPR is found in at least five different oncogenic fusion kinases, including TPR-MET, yet how TPR fusions promote activation of kinases and their oncogenic activities remains poorly understood. Here we report the crystal structure of TPR(2-142), the MET fusion partner of oncogenic TPR-MET. TPR(2-142) contains a continuous 124-residue α helix that forms an antiparallel tetramer from two leucine zipper-containing parallel coiled coils. Remarkably, single mutations cause strikingly different conformations of the coiled coil, indicating its highly dynamic nature. We further show that fusion of TPR(2-142) to the MET intracellular domain strongly and selectively stabilizes the αG helix of the MET kinase domain, and mutations of only the TPR leucine zipper residues at the junction to MET, but not other leucine zipper residues, abolish kinase activation. Together, these results provide critical insight into the TPR structure and its ability to induce dimerization and activation of fusion kinases.
- Center for Cancer and Cell Biology, Van Andel Research Institute, Grand Rapids, MI 49503, USA.
Organizational Affiliation: