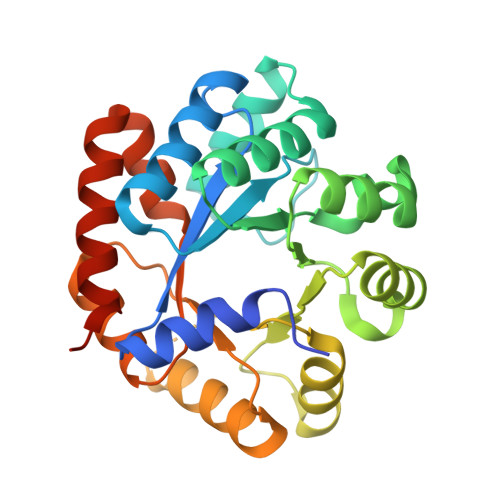
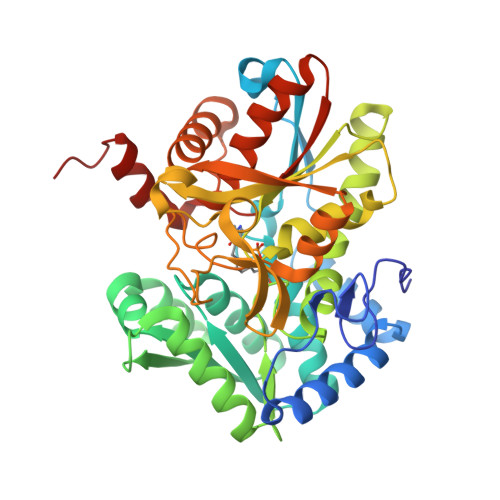
A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Wellington, S., Nag, P.P., Michalska, K., Johnston, S.E., Jedrzejczak, R.P., Kaushik, V.K., Clatworthy, A.E., Siddiqi, N., McCarren, P., Bajrami, B., Maltseva, N.I., Combs, S., Fisher, S.L., Joachimiak, A., Schreiber, S.L., Hung, D.T.(2017) Nat Chem Biol 13: 943-950
- PubMed: 28671682
- DOI: https://doi.org/10.1038/nchembio.2420
- Primary Citation of Related Structures:
5TCF, 5TCG, 5TCH, 5TCI, 5TCJ - PubMed Abstract:
New antibiotics with novel targets are greatly needed. Bacteria have numerous essential functions, but only a small fraction of such processes-primarily those involved in macromolecular synthesis-are inhibited by current drugs. Targeting metabolic enzymes has been the focus of recent interest, but effective inhibitors have been difficult to identify. We describe a synthetic azetidine derivative, BRD4592, that kills Mycobacterium tuberculosis (Mtb) through allosteric inhibition of tryptophan synthase (TrpAB), a previously untargeted, highly allosterically regulated enzyme. BRD4592 binds at the TrpAB α-β-subunit interface and affects multiple steps in the enzyme's overall reaction, resulting in inhibition not easily overcome by changes in metabolic environment. We show that TrpAB is required for the survival of Mtb and Mycobacterium marinum in vivo and that this requirement may be independent of an adaptive immune response. This work highlights the effectiveness of allosteric inhibition for targeting proteins that are naturally highly dynamic and that are essential in vivo, despite their apparent dispensability under in vitro conditions, and suggests a framework for the discovery of a next generation of allosteric inhibitors.
- The Broad Institute, Cambridge, Massachusetts, USA.
Organizational Affiliation: