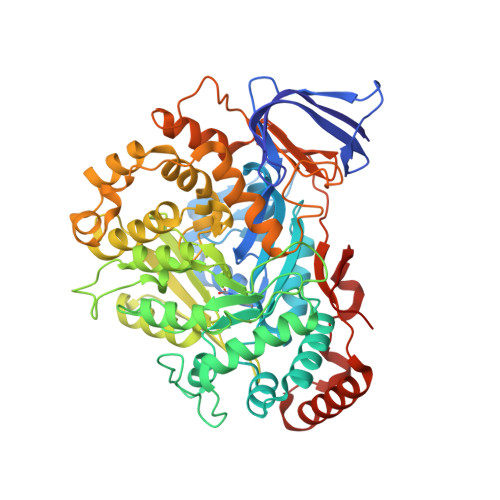
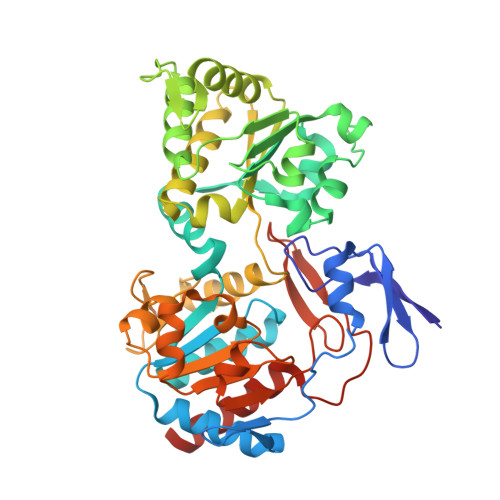
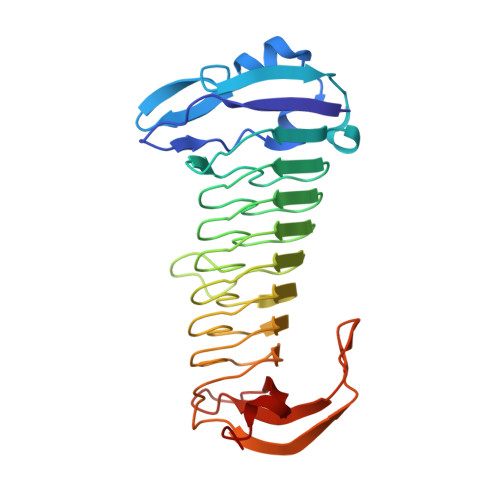
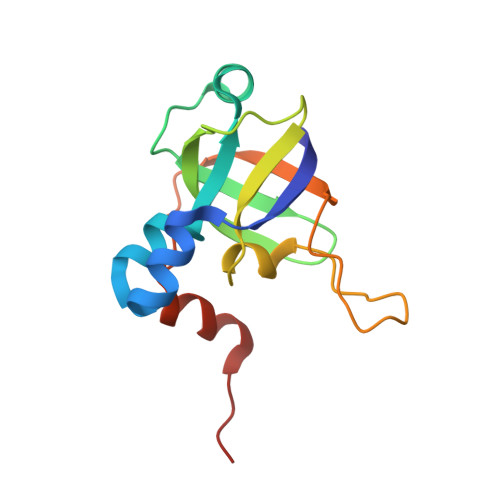
The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters.
Wagner, T., Ermler, U., Shima, S.(2016) Science 354: 114-117
- PubMed: 27846502
- DOI: https://doi.org/10.1126/science.aaf9284
- Primary Citation of Related Structures:
5T5I, 5T5M, 5T61 - PubMed Abstract:
Biological methane formation starts with a challenging adenosine triphosphate (ATP)-independent carbon dioxide (CO 2 ) fixation process. We explored this enzymatic process by solving the x-ray crystal structure of formyl-methanofuran dehydrogenase, determined here as Fwd(ABCDFG) 2 and Fwd(ABCDFG) 4 complexes, from Methanothermobacter wolfeii The latter 800-kilodalton apparatus consists of four peripheral catalytic sections and an electron-supplying core with 46 electronically coupled [4Fe-4S] clusters. Catalysis is separately performed by subunits FwdBD (FwdB and FwdD), which are related to tungsten-containing formate dehydrogenase, and subunit FwdA, a binuclear metal center carrying amidohydrolase. CO 2 is first reduced to formate in FwdBD, which then diffuses through a 43-angstrom-long tunnel to FwdA, where it condenses with methanofuran to formyl-methanofuran. The arrangement of [4Fe-4S] clusters functions as an electron relay but potentially also couples the four tungstopterin active sites over 206 angstroms.
- Max Planck Institute for Terrestrial Microbiology, Karl-von-Frisch-Straße 10, 35043 Marburg, Germany.
Organizational Affiliation: