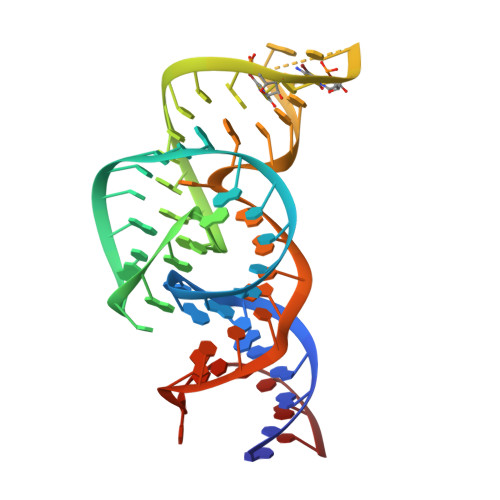
The structure of a nucleolytic ribozyme that employs a catalytic metal ion.
Liu, Y., Wilson, T.J., Lilley, D.M.(2017) Nat Chem Biol 13: 508-513
- PubMed: 28263963
- DOI: https://doi.org/10.1038/nchembio.2333
- Primary Citation of Related Structures:
5T5A - PubMed Abstract:
The TS ribozyme (originally called "twister sister") is a catalytic RNA. We present a crystal structure of the ribozyme in a pre-reactive conformation. Two co-axial helical stacks are organized by a three-way junction and two tertiary contacts. Five divalent metal ions are directly coordinated to RNA ligands, making important contributions to the RNA architecture. The scissile phosphate lies in a quasihelical loop region that is organized by a network of hydrogen bonding. A divalent metal ion is directly bound to the nucleobase 5' to the scissile phosphate, with an inner-sphere water molecule positioned to interact with the O2' nucleophile. The rate of ribozyme cleavage correlated in a log-linear manner with divalent metal ion pK a , consistent with proton transfer in the transition state, and we propose that the bound metal ion is a likely general base for the cleavage reaction. Our data indicate that the TS ribozyme functions predominantly as a metalloenzyme.
- Cancer Research UK Nucleic Acid Structure Research Group, MSI/WTB Complex, The University of Dundee, Dundee, UK.
Organizational Affiliation: