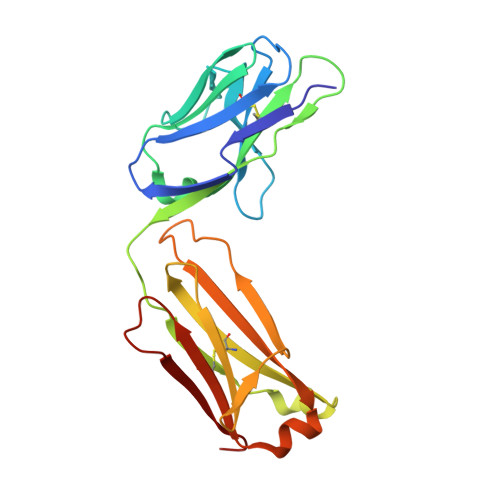
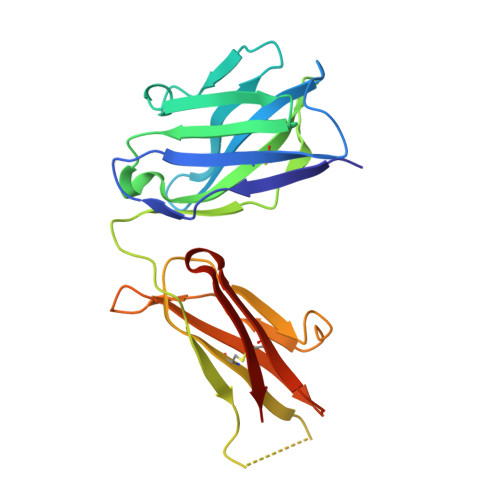
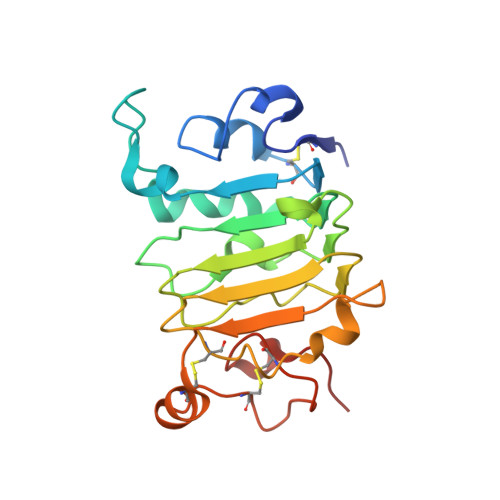
The Panitumumab EGFR Complex Reveals a Binding Mechanism That Overcomes Cetuximab Induced Resistance.
Sickmier, E.A., Kurzeja, R.J., Michelsen, K., Vazir, M., Yang, E., Tasker, A.S.(2016) PLoS One 11: e0163366-e0163366
- PubMed: 27658254
- DOI: https://doi.org/10.1371/journal.pone.0163366
- Primary Citation of Related Structures:
5SX4, 5SX5 - PubMed Abstract:
Panitumumab and cetuximab target the epidermal growth factor receptor for the treatment of metastatic colorectal cancer. These therapies provide a significant survival benefit to patients with metastatic colorectal cancer with wild-type RAS. A single point mutation in the ectodomain of EGFR (S468R) confers acquired or secondary resistance in cetuximab treated patients, which is not observed in panitumumab-treated patients. Structural and biophysical studies presented here show this mutation directly blocks cetuximab binding to EGFR domain III and describes a unique mechanism by which panitumumab uses a central cavity to accommodate this mutation.
- Depatment of Therapeutic Discovery, Amgen Inc., 360 Binney Street, Cambridge, MA 02142, United States of America.
Organizational Affiliation: