X-ray Free Electron Laser Determination of Crystal Structures of Dark and Light States of a Reversibly Photoswitching Fluorescent Protein at Room Temperature.
Hutchison, C.D.M., Cordon-Preciado, V., Morgan, R.M.L., Nakane, T., Ferreira, J., Dorlhiac, G., Sanchez-Gonzalez, A., Johnson, A.S., Fitzpatrick, A., Fare, C., Marangos, J.P., Yoon, C.H., Hunter, M.S., DePonte, D.P., Boutet, S., Owada, S., Tanaka, R., Tono, K., Iwata, S., van Thor, J.J.(2017) Int J Mol Sci 18
- PubMed: 28880248
- DOI: https://doi.org/10.3390/ijms18091918
- Primary Citation of Related Structures:
5OOZ, 5OQ9, 5OQA, 5OQE - PubMed Abstract:
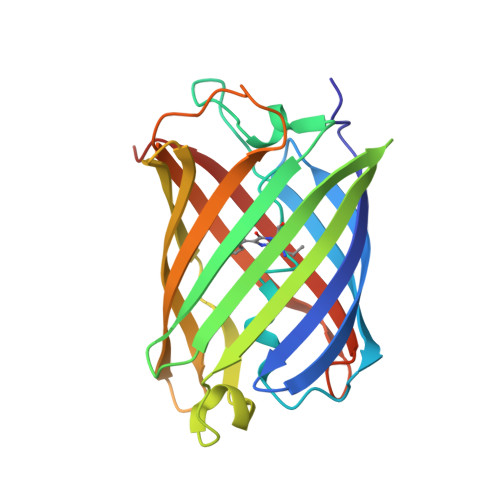
The photochromic fluorescent protein Skylan-NS (Nonlinear Structured illumination variant mEos3.1H62L) is a reversibly photoswitchable fluorescent protein which has an unilluminated/ground state with an anionic and cis chromophore conformation and high fluorescence quantum yield. Photo-conversion with illumination at 515 nm generates a meta-stable intermediate with neutral trans-chromophore structure that has a 4 h lifetime. We present X-ray crystal structures of the cis (on) state at 1.9 Angstrom resolution and the trans (off) state at a limiting resolution of 1.55 Angstrom from serial femtosecond crystallography experiments conducted at SPring-8 Angstrom Compact Free Electron Laser (SACLA) at 7.0 keV and 10.5 keV, and at Linac Coherent Light Source (LCLS) at 9.5 keV. We present a comparison of the data reduction and structure determination statistics for the two facilities which differ in flux, beam characteristics and detector technologies. Furthermore, a comparison of droplet on demand, grease injection and Gas Dynamic Virtual Nozzle (GDVN) injection shows no significant differences in limiting resolution. The photoconversion of the on- to the off-state includes both internal and surface exposed protein structural changes, occurring in regions that lack crystal contacts in the orthorhombic crystal form.
- Molecular Biophysics, Imperial College London, South Kensington Campus, London SW7 2AZ, UK. christopher.hutchison05@imperial.ac.uk.
Organizational Affiliation: