A Linear Diubiquitin-Based Probe for Efficient and Selective Detection of the Deubiquitinating Enzyme OTULIN.
Weber, A., Elliott, P.R., Pinto-Fernandez, A., Bonham, S., Kessler, B.M., Komander, D., El Oualid, F., Krappmann, D.(2017) Cell Chem Biol 24: 1299-1313.e7
- PubMed: 28919039
- DOI: https://doi.org/10.1016/j.chembiol.2017.08.006
- Primary Citation of Related Structures:
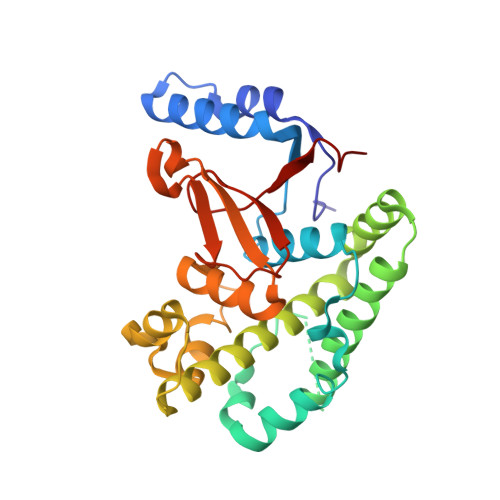
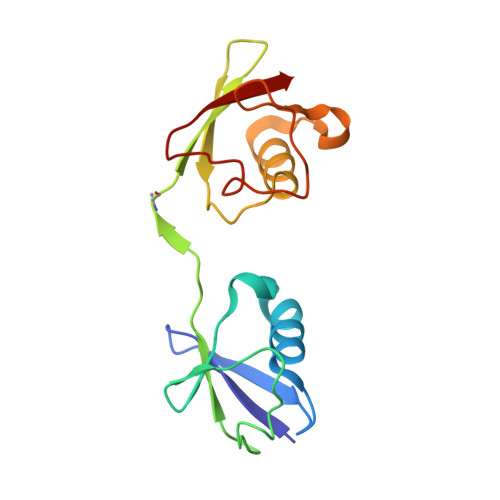
5OE7 - PubMed Abstract:
The methionine 1 (M1)-specific deubiquitinase (DUB) OTULIN acts as a negative regulator of nuclear factor κB signaling and immune homeostasis. By replacing Gly76 in distal ubiquitin (Ub) by dehydroalanine we designed the diubiquitin (diUb) activity-based probe Ub G76Dha -Ub (OTULIN activity-based probe [ABP]) that couples to the catalytic site of OTULIN and thereby captures OTULIN in its active conformation. The OTULIN ABP displays high selectivity for OTULIN and does not label other M1-cleaving DUBs, including CYLD. The only detectable cross-reactivities were the labeling of USP5 (Isopeptidase T) and an ATP-dependent assembly of polyOTULIN ABP chains via Ub-activating E1 enzymes. Both cross-reactivities were abolished by the removal of the C-terminal Gly in the ABP's proximal Ub, yielding the specific OTULIN probe Ub G76Dha -Ub ΔG76 (OTULIN ABPΔG76). Pull-downs demonstrate that substrate-bound OTULIN associates with the linear ubiquitin chain assembly complex (LUBAC). Thus, we present a highly selective ABP for OTULIN that will facilitate studying the cellular function of this essential DUB.
- Research Unit Cellular Signal Integration, Institute of Molecular Toxicology and Pharmacology, Helmholtz Zentrum München - German Research Center for Environmental Health, Ingolstaedter Landstrasse 1, 85764 Neuherberg, Germany.
Organizational Affiliation: