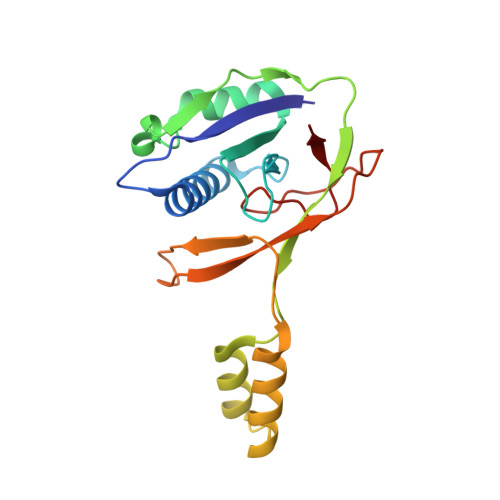
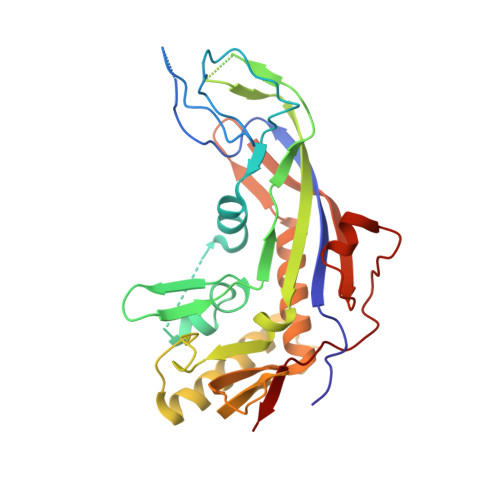
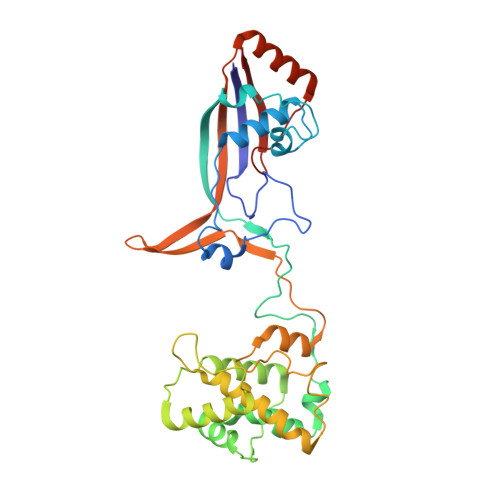
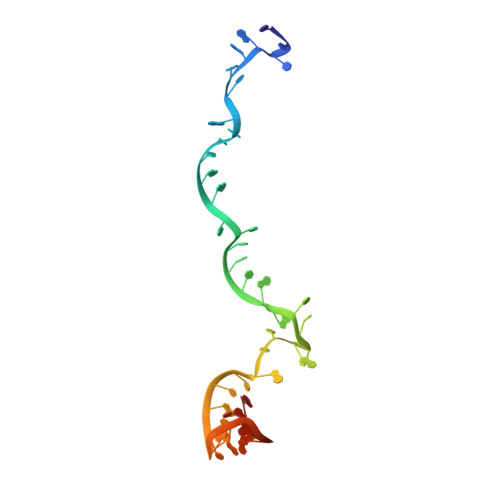
Structural Variation of Type I-F CRISPR RNA Guided DNA Surveillance.
Pausch, P., Muller-Esparza, H., Gleditzsch, D., Altegoer, F., Randau, L., Bange, G.(2017) Mol Cell 67: 622-632.e4
- PubMed: 28781236
- DOI: https://doi.org/10.1016/j.molcel.2017.06.036
- Primary Citation of Related Structures:
5O6U, 5O7H - PubMed Abstract:
CRISPR-Cas systems are prokaryotic immune systems against invading nucleic acids. Type I CRISPR-Cas systems employ highly diverse, multi-subunit surveillance Cascade complexes that facilitate duplex formation between crRNA and complementary target DNA for R-loop formation, retention, and DNA degradation by the subsequently recruited nuclease Cas3. Typically, the large subunit recognizes bona fide targets through the PAM (protospacer adjacent motif), and the small subunit guides the non-target DNA strand. Here, we present the Apo- and target-DNA-bound structures of the I-Fv (type I-F variant) Cascade lacking the small and large subunits. Large and small subunits are functionally replaced by the 5' terminal crRNA cap Cas5fv and the backbone protein Cas7fv, respectively. Cas5fv facilitates PAM recognition from the DNA major groove site, in contrast to all other described type I systems. Comparison of the type I-Fv Cascade with an anti-CRISPR protein-bound I-F Cascade reveals that the type I-Fv structure differs substantially at known anti-CRISPR protein target sites and might therefore be resistant to viral Cascade interception.
- LOEWE Center for Synthetic Microbiology (Synmikro) and Faculty of Chemistry, Philipps-University-Marburg, Hans-Meerwein-Strasse C07, 35043 Marburg, Germany.
Organizational Affiliation: