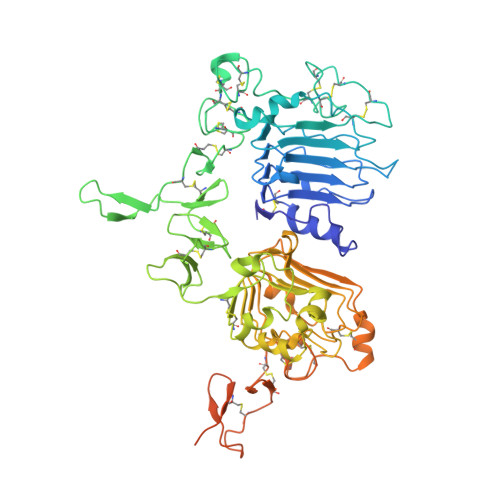
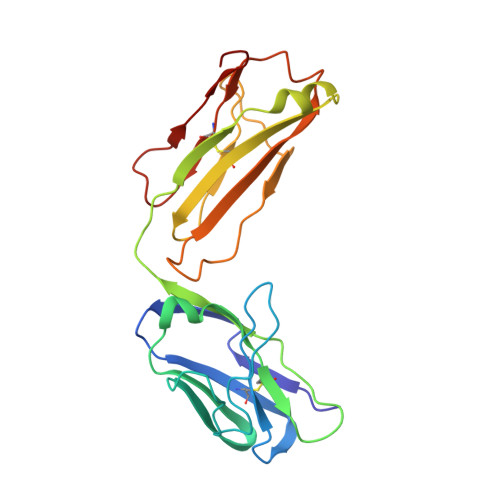
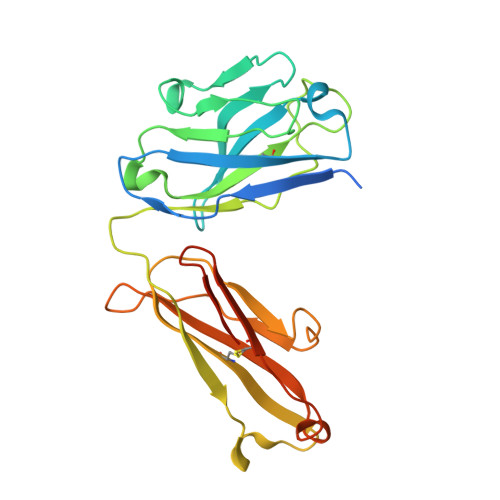
Unbiased Combinatorial Screening Identifies a Bispecific IgG1 that Potently Inhibits HER3 Signaling via HER2-Guided Ligand Blockade.
Geuijen, C.A.W., De Nardis, C., Maussang, D., Rovers, E., Gallenne, T., Hendriks, L.J.A., Visser, T., Nijhuis, R., Logtenberg, T., de Kruif, J., Gros, P., Throsby, M.(2018) Cancer Cell 33: 922-936.e10
- PubMed: 29763625
- DOI: https://doi.org/10.1016/j.ccell.2018.04.003
- Primary Citation of Related Structures:
5O4G, 5O4O, 5O7P - PubMed Abstract:
HER2-driven cancers require phosphatidylinositide-3 kinase (PI3K)/Akt signaling through HER3 to promote tumor growth and survival. The therapeutic benefit of HER2-targeting agents, which depend on PI3K/Akt inhibition, can be overcome by hyperactivation of the heregulin (HRG)/HER3 pathway. Here we describe an unbiased phenotypic combinatorial screening approach to identify a bispecific immunoglobulin G1 (IgG1) antibody against HER2 and HER3. In tumor models resistant to HER2-targeting agents, the bispecific IgG1 potently inhibits the HRG/HER3 pathway and downstream PI3K/Akt signaling via a "dock & block" mechanism. This bispecific IgG1 is a potentially effective therapy for breast cancer and other tumors with hyperactivated HRG/HER3 signaling.
- Merus NV, 3584 Utrecht, the Netherlands.
Organizational Affiliation: