Unravelling the Molecular Basis of High Affinity Nanobodies against HIV p24: In Vitro Functional, Structural, and in Silico Insights.
Gray, E.R., Brookes, J.C., Caillat, C., Turbe, V., Webb, B.L.J., Granger, L.A., Miller, B.S., McCoy, L.E., El Khattabi, M., Verrips, C.T., Weiss, R.A., Duffy, D.M., Weissenhorn, W., McKendry, R.A.(2017) ACS Infect Dis 3: 479-491
- PubMed: 28591513
- DOI: https://doi.org/10.1021/acsinfecdis.6b00189
- Primary Citation of Related Structures:
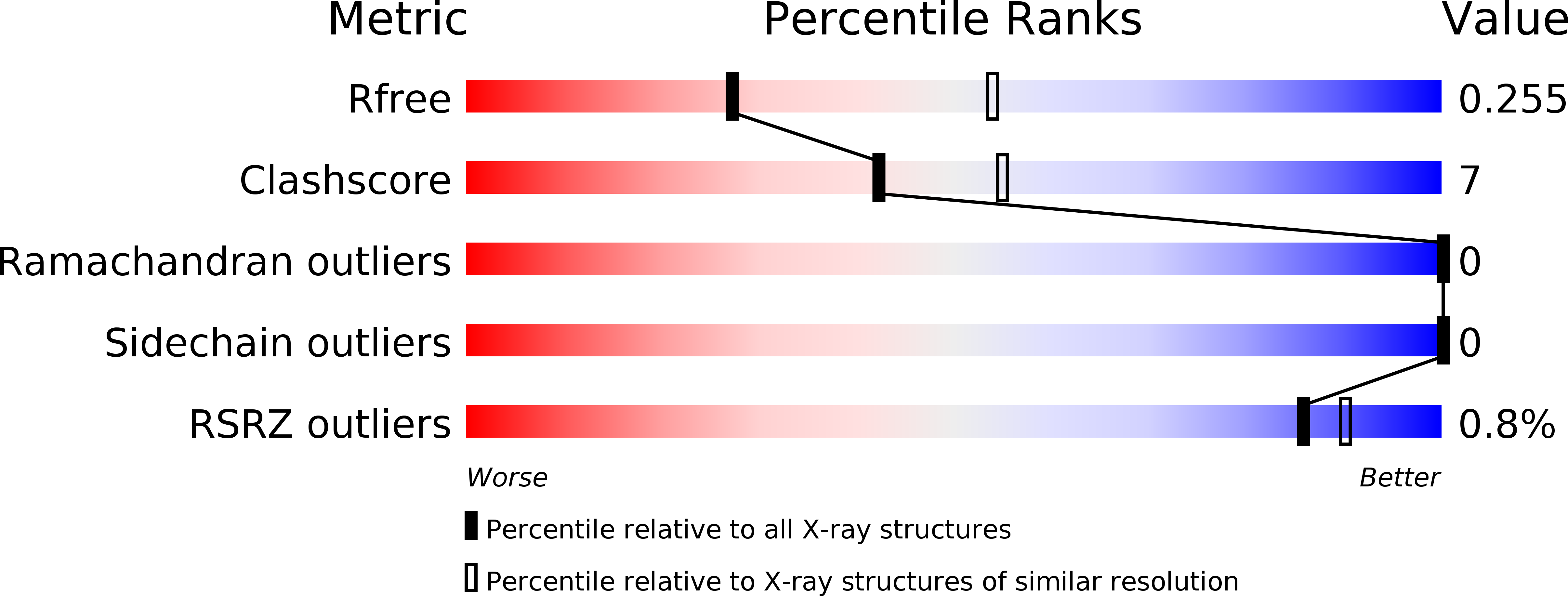
5O2U - PubMed Abstract:
Preventing the spread of infectious diseases remains an urgent priority worldwide, and this is driving the development of advanced nanotechnology to diagnose infections at the point of care. Herein, we report the creation of a library of novel nanobody capture ligands to detect p24, one of the earliest markers of HIV infection. We demonstrate that these nanobodies, one tenth the size of conventional antibodies, exhibit high sensitivity and broad specificity to global HIV-1 subtypes. Biophysical characterization indicates strong 690 pM binding constants and fast kinetic on-rates, 1 to 2 orders of magnitude better than monoclonal antibody comparators. A crystal structure of the lead nanobody and p24 was obtained and used alongside molecular dynamics simulations to elucidate the molecular basis of these enhanced performance characteristics. They indicate that binding occurs at C-terminal helices 10 and 11 of p24, a negatively charged region of p24 complemented by the positive surface of the nanobody binding interface involving CDR1, CDR2, and CDR3 loops. Our findings have broad implications on the design of novel antibodies and a wide range of advanced biomedical applications.
Organizational Affiliation:
London Centre for Nanotechnology, Division of Medicine and Department of Physics and Astronomy, University College London , 17-19 Gordon Street, London, WC1H 0AH, United Kingdom.