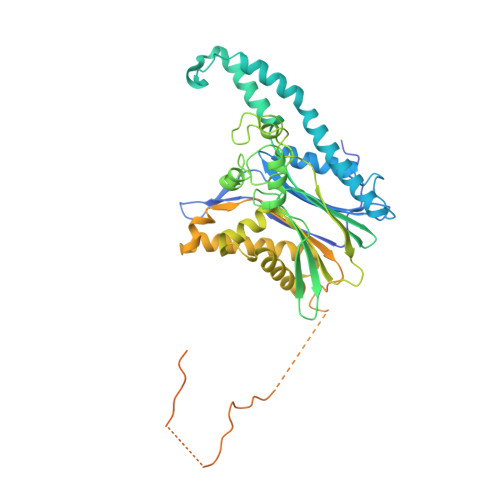
The TAB1-p38 alpha complex aggravates myocardial injury and can be targeted by small molecules.
De Nicola, G.F., Bassi, R., Nichols, C., Fernandez-Caggiano, M., Golforoush, P.A., Thapa, D., Anderson, R., Martin, E.D., Verma, S., Kleinjung, J., Laing, A., Hutchinson, J.P., Eaton, P., Clark, J., Marber, M.S.(2018) JCI Insight 3
- PubMed: 30135318
- DOI: https://doi.org/10.1172/jci.insight.121144
- Primary Citation of Related Structures:
5NZZ - PubMed Abstract:
Inhibiting MAPK14 (p38α) diminishes cardiac damage in myocardial ischemia. During myocardial ischemia, p38α interacts with TAB1, a scaffold protein, which promotes p38α autoactivation; active p38α (pp38α) then transphosphorylates TAB1. Previously, we solved the X-ray structure of the p38α-TAB1 (residues 384-412) complex. Here, we further characterize the interaction by solving the structure of the pp38α-TAB1 (residues 1-438) complex in the active state. Based on this information, we created a global knock-in (KI) mouse with substitution of 4 residues on TAB1 that we show are required for docking onto p38α. Whereas ablating p38α or TAB1 resulted in early embryonal lethality, the TAB1-KI mice were viable and had no appreciable alteration in their lymphocyte repertoire or myocardial transcriptional profile; nonetheless, following in vivo regional myocardial ischemia, infarction volume was significantly reduced and the transphosphorylation of TAB1 was disabled. Unexpectedly, the activation of myocardial p38α during ischemia was only mildly attenuated in TAB1-KI hearts. We also identified a group of fragments able to disrupt the interaction between p38α and TAB1. We conclude that the interaction between the 2 proteins can be targeted with small molecules. The data reveal that it is possible to selectively inhibit signaling downstream of p38α to attenuate ischemic injury.
- British Heart Foundation Centre of Excellence, The Rayne Institute, St. Thomas' Hospital, and.
Organizational Affiliation: