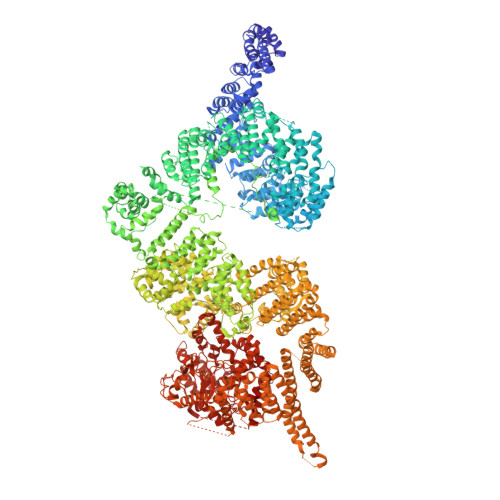
Structures of closed and open conformations of dimeric human ATM.
Baretic, D., Pollard, H.K., Fisher, D.I., Johnson, C.M., Santhanam, B., Truman, C.M., Kouba, T., Fersht, A.R., Phillips, C., Williams, R.L.(2017) Sci Adv 3: e1700933-e1700933
- PubMed: 28508083
- DOI: https://doi.org/10.1126/sciadv.1700933
- Primary Citation of Related Structures:
5NP0, 5NP1 - PubMed Abstract:
ATM (ataxia-telangiectasia mutated) is a phosphatidylinositol 3-kinase-related protein kinase (PIKK) best known for its role in DNA damage response. ATM also functions in oxidative stress response, insulin signaling, and neurogenesis. Our electron cryomicroscopy (cryo-EM) suggests that human ATM is in a dynamic equilibrium between closed and open dimers. In the closed state, the PIKK regulatory domain blocks the peptide substrate-binding site, suggesting that this conformation may represent an inactive or basally active enzyme. The active site is held in this closed conformation by interaction with a long helical hairpin in the TRD3 (tetratricopeptide repeats domain 3) domain of the symmetry-related molecule. The open dimer has two protomers with only a limited contact interface, and it lacks the intermolecular interactions that block the peptide-binding site in the closed dimer. This suggests that the open conformation may be more active. The ATM structure shows the detailed topology of the regulator-interacting N-terminal helical solenoid. The ATM conformational dynamics shown by the structures represent an important step in understanding the enzyme regulation.
- Medical Research Council Laboratory of Molecular Biology, Cambridge CB2 0QH, UK.
Organizational Affiliation: