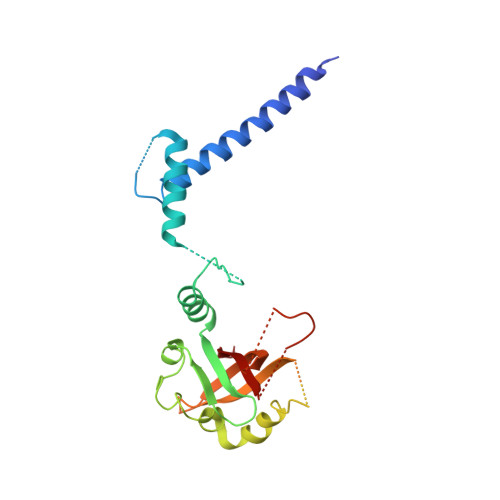
Structural Basis for Aryl Hydrocarbon Receptor-Mediated Gene Activation.
Schulte, K.W., Green, E., Wilz, A., Platten, M., Daumke, O.(2017) Structure 25: 1025-1033.e3
- PubMed: 28602820
- DOI: https://doi.org/10.1016/j.str.2017.05.008
- Primary Citation of Related Structures:
5NJ8 - PubMed Abstract:
The aryl hydrocarbon receptor (AHR) and the AHR nuclear translocator (ARNT) constitute a heterodimeric basic helix-loop-helix-Per-ARNT-Sim (bHLH-PAS) domain containing transcription factor with central functions in development and cellular homeostasis. AHR is activated by xenobiotics, notably dioxin, as well as by exogenous and endogenous metabolites. Modulation of AHR activity holds promise for the treatment of diseases featuring altered cellular homeostasis, such as cancer or autoimmune disorders. Here, we present the crystal structure of a heterodimeric AHR:ARNT complex containing the PAS A and bHLH domain bound to its target DNA. The structure provides insights into the DNA binding mode of AHR and elucidates how stable dimerization of AHR:ARNT is achieved through sophisticated domain interplay via three specific interfaces. Using mutational analyses, we prove the relevance of the observed interfaces for AHR-mediated gene activation. Thus, our work establishes the structural basis of AHR assembly and DNA interaction and provides a template for targeted drug design.
- Crystallography Department, Max-Delbrück-Center for Molecular Medicine, Robert-Rössle-Strasse 10, 13125 Berlin, Germany; Institute of Chemistry and Biochemistry, Freie Universität Berlin, Takustrasse 6, 14195 Berlin, Germany.
Organizational Affiliation: