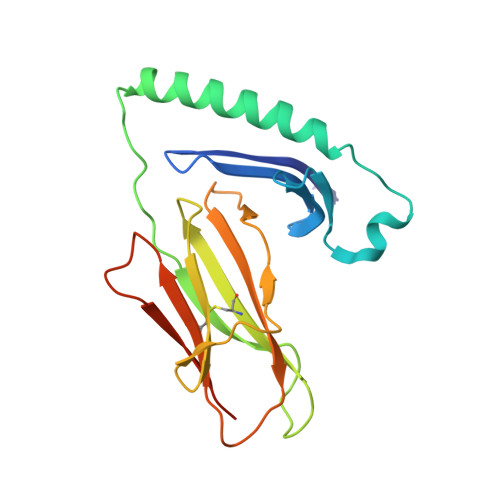
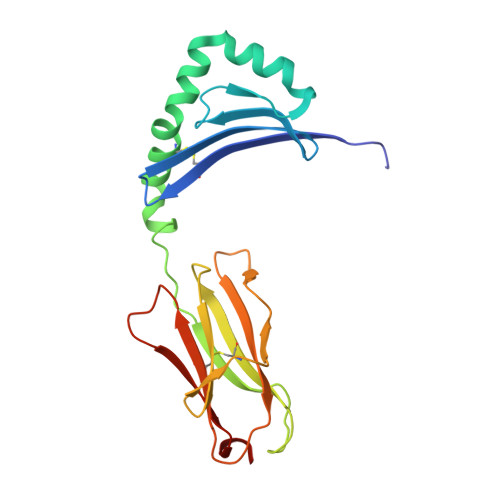
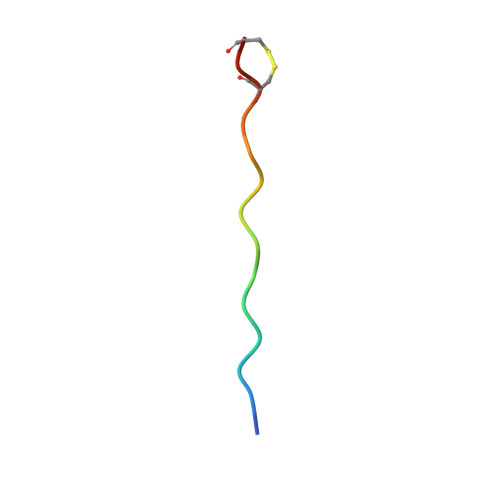
Memory T cells specific to citrullinated alpha-enolase are enriched in the rheumatic joint
Pieper, J., Dubnovitsky, A., Gerstner, C., James, E.A., Rieck, M., Kozhukh, G., Tandre, K., Pellegrino, S., Gebe, J.A., Ronnblom, L., Sandalova, T., Kwok, W.W., Klareskog, L., Buckner, J.H., Achour, A., Malmstrom, V.(2018) J Autoimmun 92: 47-56
- PubMed: 29853344
- DOI: https://doi.org/10.1016/j.jaut.2018.04.004
- Primary Citation of Related Structures:
5NI9, 5NIG - PubMed Abstract:
ACPA-positive rheumatoid arthritis (RA) is associated with distinct HLA-DR alleles and immune responses to many citrullinated self-antigens. Herein we investigated the T cell epitope confined within α-enolase 326-340 in the context of HLA-DRB1*04:01 and assessed the corresponding CD4 + T cells in both the circulation and in the rheumatic joint. Comparative crystallographic analyses were performed for the native and citrullinated α-enolase 326-340 peptides in complex with HLA-DRB1*04:01. HLA-tetramers assembled with either the native or citrullinated peptide were used for ex vivo and in vitro assessment of α-enolase-specific T cells in peripheral blood, synovial fluid and synovial tissue by flow cytometry. The native and modified peptides take a completely conserved structural conformation within the peptide-binding cleft of HLA-DRB1*04:01. The citrulline residue-327 was located N-terminally, protruding towards TCRs. The frequencies of T cells recognizing native eno 326-340 were similar in synovial fluid and peripheral blood, while in contrast, the frequency of T cells recognizing cit-eno 326-340 was significantly elevated in synovial fluid compared to peripheral blood (3.6-fold, p = 0.0150). Additionally, citrulline-specific T cells with a memory phenotype were also significantly increased (1.6-fold, p = 0.0052) in synovial fluid compared to peripheral blood. The native T cell epitope confined within α-enolase 326-340 does not appear to lead to complete negative selection of cognate CD4 + T cells. In RA patient samples, only T cells recognizing the citrullinated version of α-enolase 326-340 were found at elevated frequencies implicating that neo-antigen formation is critical for breach of tolerance.
- Rheumatology Unit, Department of Medicine Solna, Center for Molecular Medicine, Karolinska Institutet, Karolinska University Hospital, Stockholm, Sweden.
Organizational Affiliation: