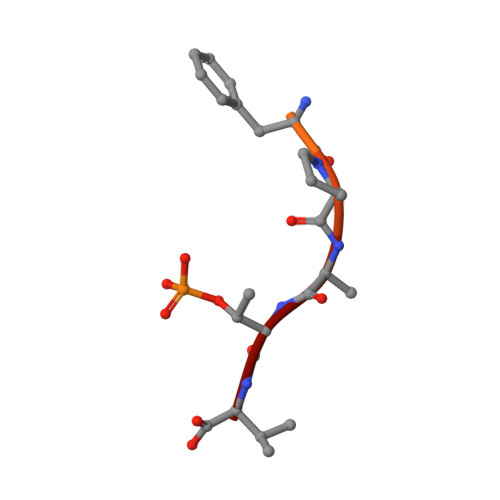
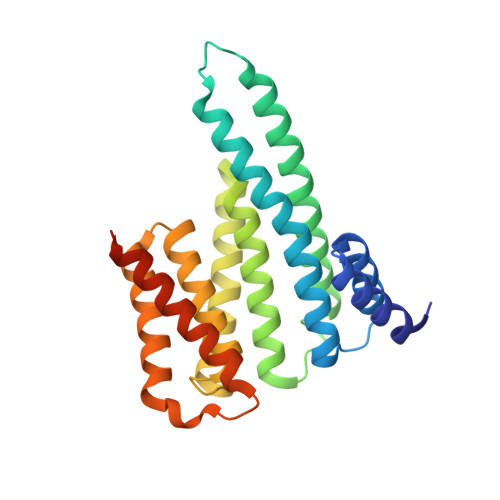
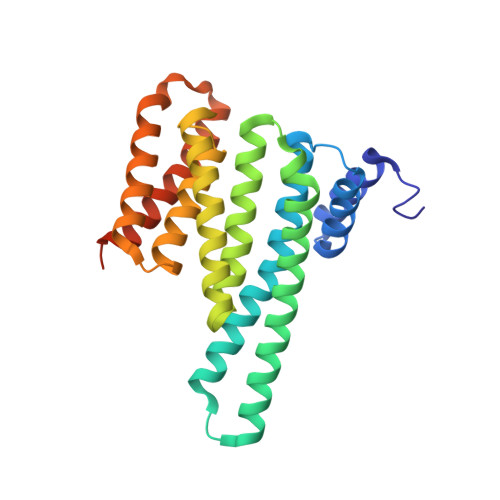
A Binary Bivalent Supramolecular Assembly Platform Based on Cucurbit[8]uril and Dimeric Adapter Protein 14-3-3.
de Vink, P.J., Briels, J.M., Schrader, T., Milroy, L.G., Brunsveld, L., Ottmann, C.(2017) Angew Chem Int Ed Engl 56: 8998-9002
- PubMed: 28510303
- DOI: https://doi.org/10.1002/anie.201701807
- Primary Citation of Related Structures:
5N10 - PubMed Abstract:
Interactions between proteins frequently involve recognition sequences based on multivalent binding events. Dimeric 14-3-3 adapter proteins are a prominent example and typically bind partner proteins in a phosphorylation-dependent mono- or bivalent manner. Herein we describe the development of a cucurbit[8]uril (Q8)-based supramolecular system, which in conjunction with the 14-3-3 protein dimer acts as a binary and bivalent protein assembly platform. We fused the phenylalanine-glycine-glycine (FGG) tripeptide motif to the N-terminus of the 14-3-3-binding epitope of the estrogen receptor α (ERα) for selective binding to Q8. Q8-induced dimerization of the ERα epitope augmented its affinity towards 14-3-3 through a binary bivalent binding mode. The crystal structure of the Q8-induced ternary complex revealed molecular insight into the multiple supramolecular interactions between the protein, the peptide, and Q8.
- Laboratory of Chemical Biology and Institute of Complex Molecular Systems, Department of Biomedical Engineering, Eindhoven University of Technology, Den Dolech 2, 5612 AZ, Eindhoven, The Netherlands.
Organizational Affiliation: