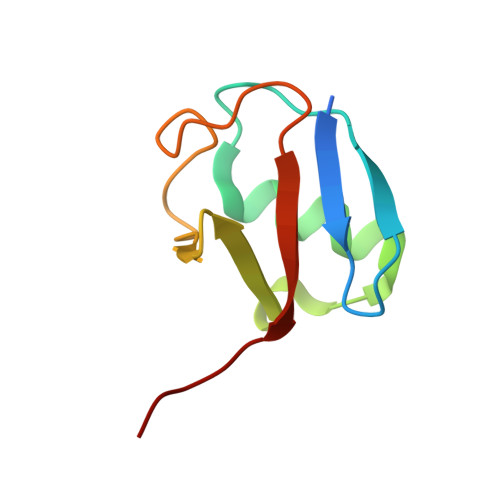
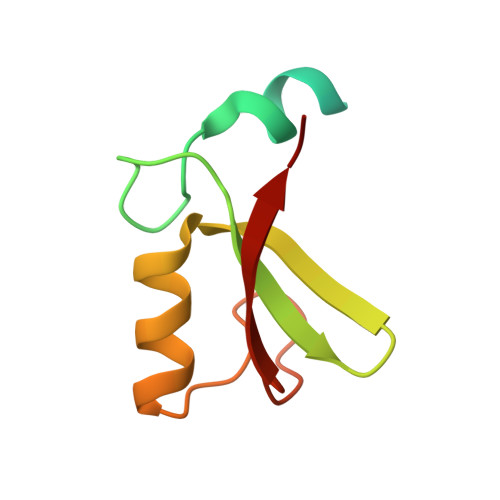
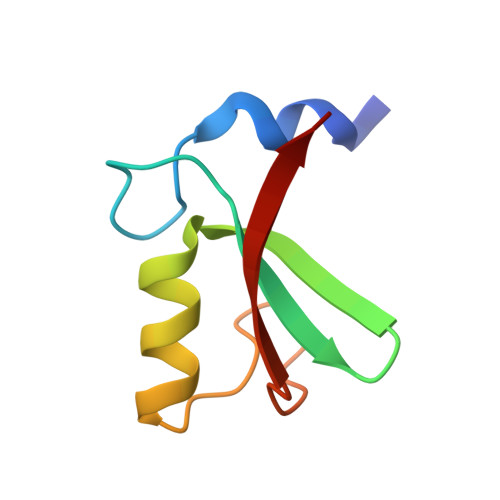
Structural analysis of MDM2 RING separates degradation from regulation of p53 transcription activity.
Nomura, K., Klejnot, M., Kowalczyk, D., Hock, A.K., Sibbet, G.J., Vousden, K.H., Huang, D.T.(2017) Nat Struct Mol Biol 24: 578-587
- PubMed: 28553961
- DOI: https://doi.org/10.1038/nsmb.3414
- Primary Citation of Related Structures:
5MNJ - PubMed Abstract:
MDM2-MDMX complexes bind the p53 tumor-suppressor protein, inhibiting p53's transcriptional activity and targeting p53 for proteasomal degradation. Inhibitors that disrupt binding between p53 and MDM2 efficiently activate a p53 response, but their use in the treatment of cancers that retain wild-type p53 may be limited by on-target toxicities due to p53 activation in normal tissue. Guided by a novel crystal structure of the MDM2-MDMX-E2(UbcH5B)-ubiquitin complex, we designed MDM2 mutants that prevent E2-ubiquitin binding without altering the RING-domain structure. These mutants lack MDM2's E3 activity but retain the ability to limit p53's transcriptional activity and allow cell proliferation. Cells expressing these mutants respond more quickly to cellular stress than cells expressing wild-type MDM2, but basal p53 control is maintained. Targeting the MDM2 E3-ligase activity could therefore widen the therapeutic window of p53 activation in tumors.
- Cancer Research-UK Beatson Institute, Garscube Estate, Glasgow, UK.
Organizational Affiliation: