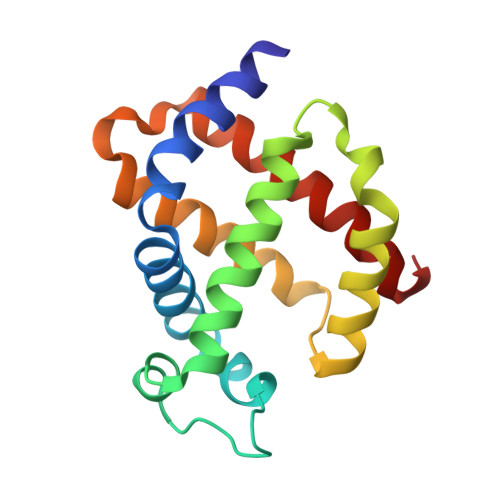
Ligand pathways in neuroglobin revealed by low-temperature photodissociation and docking experiments.
Ardiccioni, C., Arcovito, A., Della Longa, S., van der Linden, P., Bourgeois, D., Weik, M., Montemiglio, L.C., Savino, C., Avella, G., Exertier, C., Carpentier, P., Prange, T., Brunori, M., Colloc'h, N., Vallone, B.(2019) IUCrJ 6: 832-842
- PubMed: 31576217
- DOI: https://doi.org/10.1107/S2052252519008157
- Primary Citation of Related Structures:
5MJC, 5MJD, 6I3T, 6I40 - PubMed Abstract:
A combined biophysical approach was applied to map gas-docking sites within murine neuroglobin (Ngb), revealing snapshots of events that might govern activity and dynamics in this unique hexacoordinate globin, which is most likely to be involved in gas-sensing in the central nervous system and for which a precise mechanism of action remains to be elucidated. The application of UV-visible microspectroscopy in crystallo , solution X-ray absorption near-edge spectroscopy and X-ray diffraction experiments at 15-40 K provided the structural characterization of an Ngb photolytic intermediate by cryo-trapping and allowed direct observation of the relocation of carbon monoxide within the distal heme pocket after photodissociation. Moreover, X-ray diffraction at 100 K under a high pressure of dioxygen, a physiological ligand of Ngb, unravelled the existence of a storage site for O 2 in Ngb which coincides with Xe-III, a previously described docking site for xenon or krypton. Notably, no other secondary sites were observed under our experimental conditions.
- Department of Life and Environmental Sciences, New York-Marche Structural Biology Center (NY-MaSBiC), Polytechnic University of Marche, Ancona, Italy.
Organizational Affiliation: