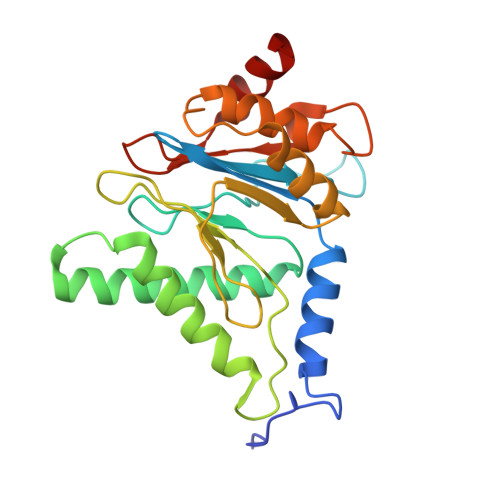
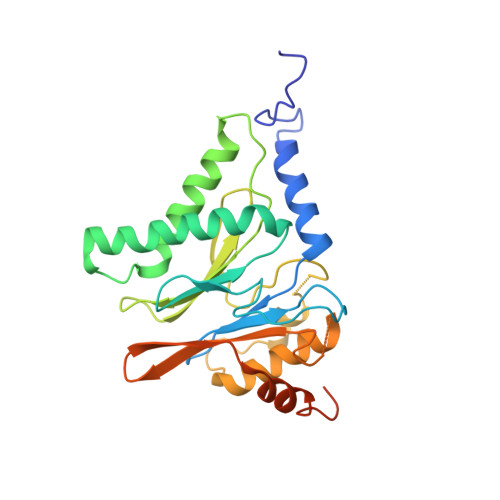
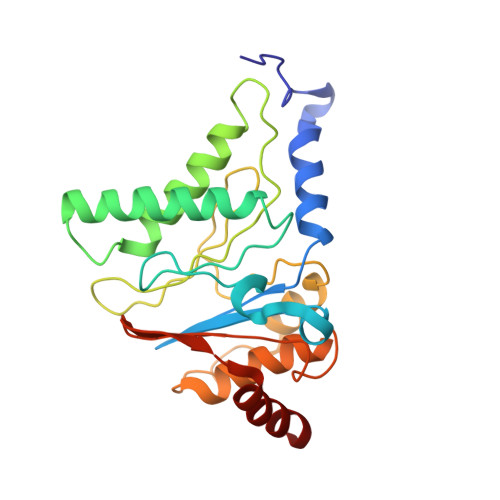
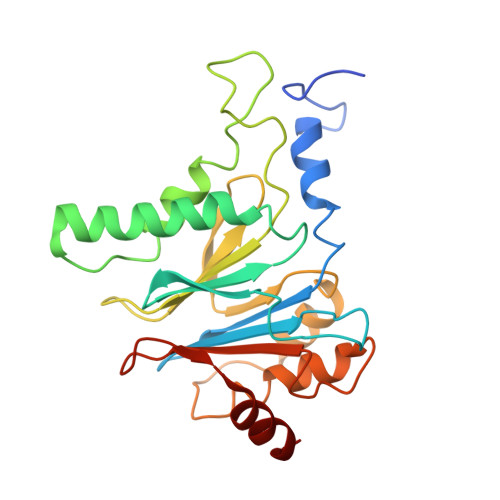
Long-range allosteric regulation of the human 26S proteasome by 20S proteasome-targeting cancer drugs.
Haselbach, D., Schrader, J., Lambrecht, F., Henneberg, F., Chari, A., Stark, H.(2017) Nat Commun 8: 15578-15578
- PubMed: 28541292
- DOI: https://doi.org/10.1038/ncomms15578
- Primary Citation of Related Structures:
5M32 - PubMed Abstract:
The proteasome holoenzyme is the major non-lysosomal protease; its proteolytic activity is essential for cellular homeostasis. Thus, it is an attractive target for the development of chemotherapeutics. While the structural basis of core particle (CP) inhibitors is largely understood, their structural impact on the proteasome holoenzyme remains entirely elusive. Here, we determined the structure of the 26S proteasome with and without the inhibitor Oprozomib. Drug binding modifies the energy landscape of conformational motion in the proteasome regulatory particle (RP). Structurally, the energy barrier created by Oprozomib triggers a long-range allosteric regulation, resulting in the stabilization of a non-productive state. Thereby, the chemical drug-binding signal is converted, propagated and amplified into structural changes over a distance of more than 150 Å from the proteolytic site to the ubiquitin receptor Rpn10. The direct visualization of changes in conformational dynamics upon drug binding allows new ways to screen and develop future allosteric proteasome inhibitors.
- Department for Structural Dynamics, Max-Planck Institute for Biophysical Chemistry, Am Fassberg 11, 37077 Göttingen, Germany.
Organizational Affiliation: