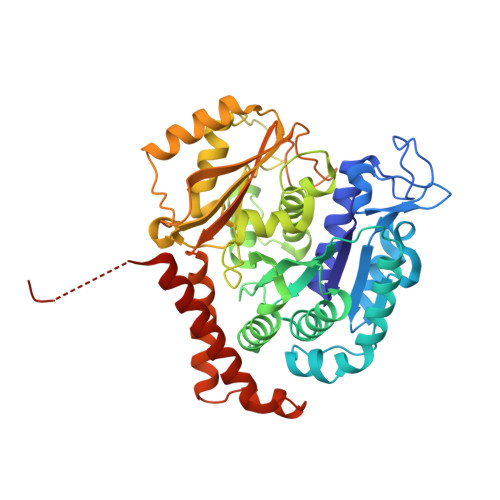
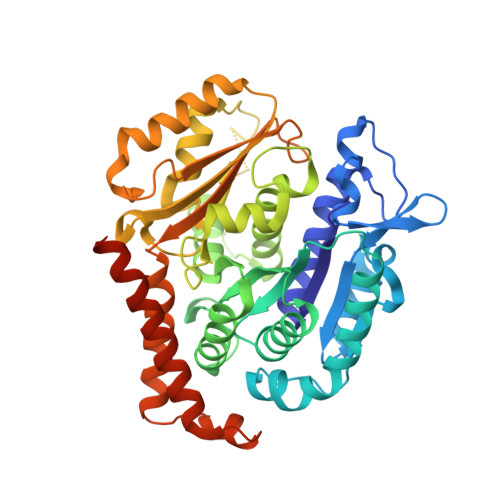
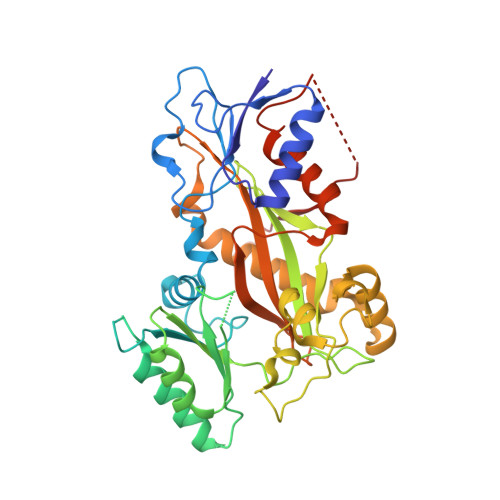
Structural Basis of Microtubule Stabilization by Discodermolide.
Prota, A.E., Bargsten, K., Redondo-Horcajo, M., Smith, A.B., Yang, C.H., McDaid, H.M., Paterson, I., Horwitz, S.B., Fernando Diaz, J., Steinmetz, M.O.(2017) Chembiochem 18: 905-909
- PubMed: 28207984
- DOI: https://doi.org/10.1002/cbic.201600696
- Primary Citation of Related Structures:
5LXS, 5LXT - PubMed Abstract:
Microtubule-stabilizing agents (MSAs) are widely used in chemotherapy. Using X-ray crystallography we elucidated the detailed binding modes of two potent MSAs, (+)-discodermolide (DDM) and the DDM-paclitaxel hybrid KS-1-199-32, in the taxane pocket of β-tubulin. The two compounds bind in a very similar hairpin conformation, as previously observed in solution. However, they stabilize the M-loop of β-tubulin differently: KS-1-199-32 induces an M-loop helical conformation that is not observed for DDM. In the context of the microtubule structure, both MSAs connect the β-tubulin helices H6 and H7 and loop S9-S10 with the M-loop. This is similar to the structural effects elicited by epothilone A, but distinct from paclitaxel. Together, our data reveal differential binding mechanisms of DDM and KS-1-199-32 on tubulin.
- Laboratory of Biomolecular Research, Department of Biology and Chemistry, Paul Scherrer Institut, OFLC/111, 5232, Villigen PSI, Switzerland.
Organizational Affiliation: