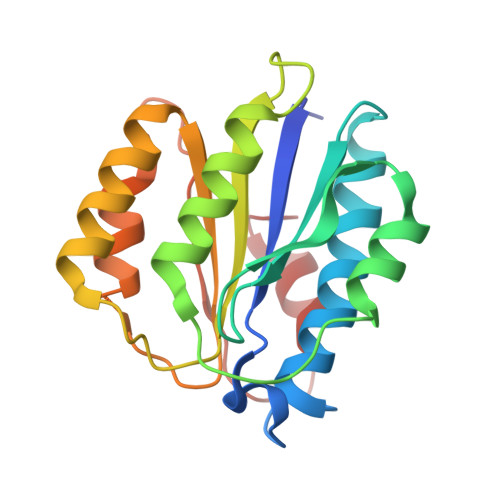
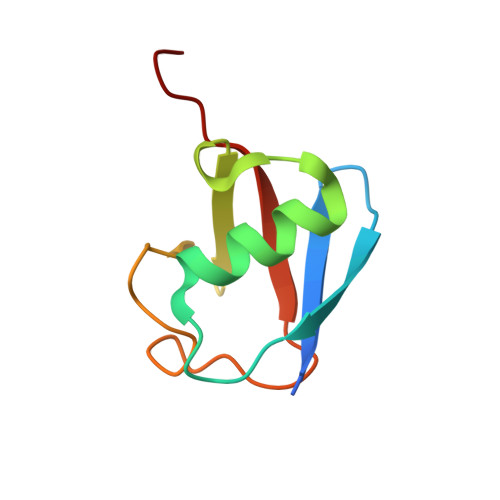
Structure of ubiquitylated-Rpn10 provides insight into its autoregulation mechanism.
Keren-Kaplan, T., Zeev Peters, L., Levin-Kravets, O., Attali, I., Kleifeld, O., Shohat, N., Artzi, S., Zucker, O., Pilzer, I., Reis, N., Glickman, M.H., Ben-Aroya, S., Prag, G.(2016) Nat Commun 7: 12960-12960
- PubMed: 27698474
- DOI: https://doi.org/10.1038/ncomms12960
- Primary Citation of Related Structures:
5LN1 - PubMed Abstract:
Ubiquitin receptors decode ubiquitin signals into many cellular responses. Ubiquitin receptors also undergo coupled monoubiquitylation, and rapid deubiquitylation has hampered the characterization of the ubiquitylated state. Using bacteria that express a ubiquitylation apparatus, we purified and determined the crystal structure of the proteasomal ubiquitin-receptor Rpn10 in its ubiquitylated state. The structure shows a novel ubiquitin-binding patch that directs K84 ubiquitylation. Superimposition of ubiquitylated-Rpn10 onto electron-microscopy models of proteasomes indicates that the Rpn10-conjugated ubiquitin clashes with Rpn9, suggesting that ubiquitylation might be involved in releasing Rpn10 from the proteasome. Indeed, ubiquitylation on immobilized proteasomes dissociates the modified Rpn10 from the complex, while unmodified Rpn10 mainly remains associated. In vivo experiments indicate that contrary to wild type, Rpn10-K84R is stably associated with the proteasomal subunit Rpn9. Similarly Rpn10, but not ubiquitylated-Rpn10, binds Rpn9 in vitro. Thus we suggest that ubiquitylation functions to dissociate modified ubiquitin receptors from their targets, a function that promotes cyclic activity of ubiquitin receptors.
- Department of Biochemistry and Molecular Biology and the Institute of Structural Biology, George S. Wise Faculty of Life Sciences, Tel Aviv University, Tel Aviv 69978, Israel.
Organizational Affiliation: