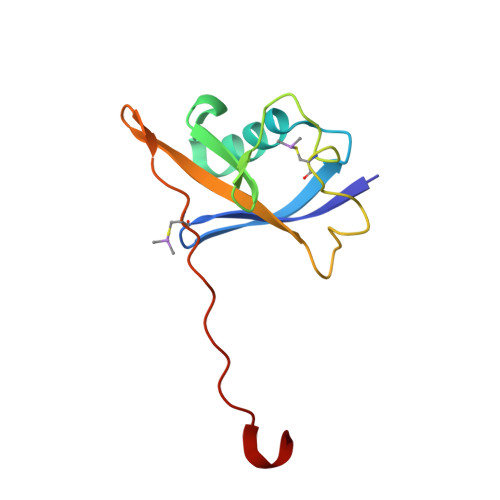
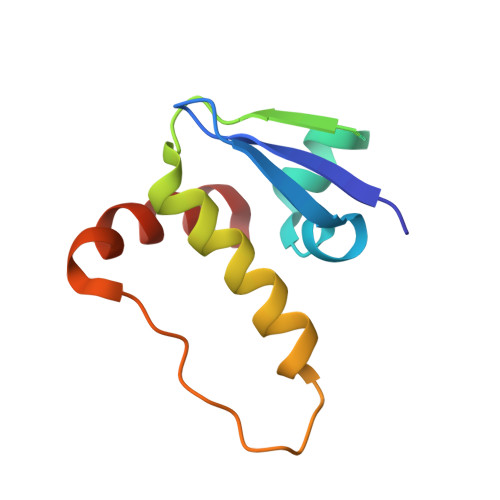
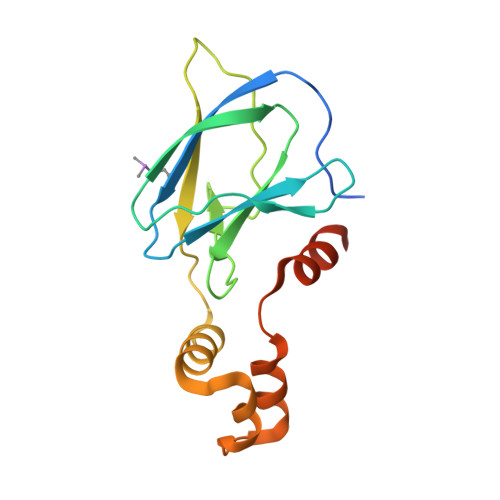
Potent and selective chemical probe of hypoxic signalling downstream of HIF-alpha hydroxylation via VHL inhibition.
Frost, J., Galdeano, C., Soares, P., Gadd, M.S., Grzes, K.M., Ellis, L., Epemolu, O., Shimamura, S., Bantscheff, M., Grandi, P., Read, K.D., Cantrell, D.A., Rocha, S., Ciulli, A.(2016) Nat Commun 7: 13312-13312
- PubMed: 27811928
- DOI: https://doi.org/10.1038/ncomms13312
- Primary Citation of Related Structures:
5LLI - PubMed Abstract:
Chemical strategies to using small molecules to stimulate hypoxia inducible factors (HIFs) activity and trigger a hypoxic response under normoxic conditions, such as iron chelators and inhibitors of prolyl hydroxylase domain (PHD) enzymes, have broad-spectrum activities and off-target effects. Here we disclose VH298, a potent VHL inhibitor that stabilizes HIF-α and elicits a hypoxic response via a different mechanism, that is the blockade of the VHL:HIF-α protein-protein interaction downstream of HIF-α hydroxylation by PHD enzymes. We show that VH298 engages with high affinity and specificity with VHL as its only major cellular target, leading to selective on-target accumulation of hydroxylated HIF-α in a concentration- and time-dependent fashion in different cell lines, with subsequent upregulation of HIF-target genes at both mRNA and protein levels. VH298 represents a high-quality chemical probe of the HIF signalling cascade and an attractive starting point to the development of potential new therapeutics targeting hypoxia signalling.
- Division of Biological Chemistry and Drug Discovery, School of Life Sciences, University of Dundee, Dow Street, Dundee DD1 5EH, Scotland, UK.
Organizational Affiliation: