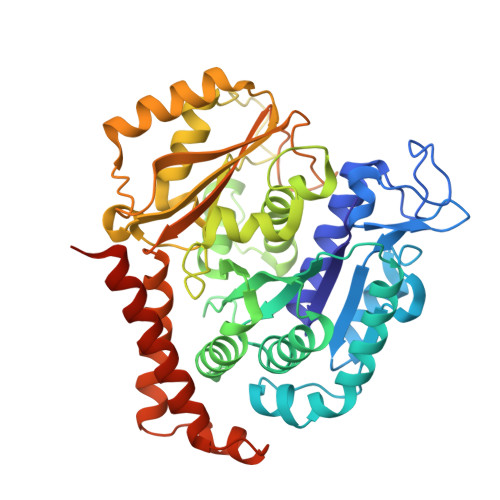
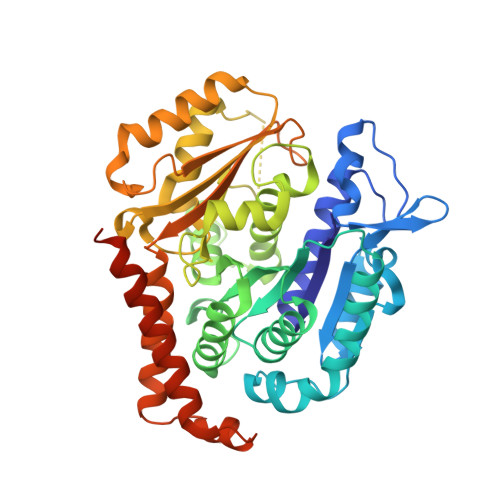
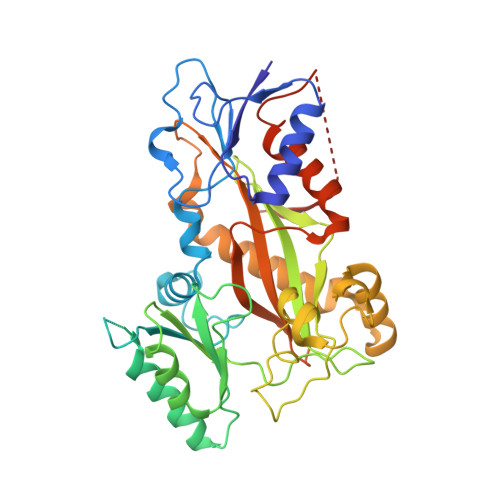
Pironetin Binds Covalently to alpha Cys316 and Perturbs a Major Loop and Helix of alpha-Tubulin to Inhibit Microtubule Formation.
Prota, A.E., Setter, J., Waight, A.B., Bargsten, K., Murga, J., Diaz, J.F., Steinmetz, M.O.(2016) J Mol Biology 428: 2981-2988
- PubMed: 27395016
- DOI: https://doi.org/10.1016/j.jmb.2016.06.023
- Primary Citation of Related Structures:
5LA6 - PubMed Abstract:
Microtubule-targeting agents are among the most powerful drugs used in chemotherapy to treat cancer patients. Pironetin is a natural product that displays promising anticancer properties by binding to and potently inhibiting tubulin assembly into microtubules; however, its molecular mechanism of action remained obscure. Here, we solved the crystal structure of the tubulin-pironetin complex and found that the compound covalently binds to Cys316 of α-tubulin. The structure further revealed that pironetin perturbs the T7 loop and helix H8 of α-tubulin. Since both these elements are essential for establishing longitudinal tubulin contacts in microtubules, this result explains how pironetin inhibits the formation of microtubules. Together, our data define the molecular details of the pironetin binding site on α-tubulin and thus offer a promising basis for the rational design of pironetin variants with improved activity profiles. They further extend our knowledge on strategies evolved by natural products to target and perturb the microtubule cytoskeleton.
- Laboratory of Biomolecular Research, Department of Biology and Chemistry, Paul Scherrer Institut, 5232 Villigen, Switzerland. Electronic address: andrea.prota@psi.ch.
Organizational Affiliation: