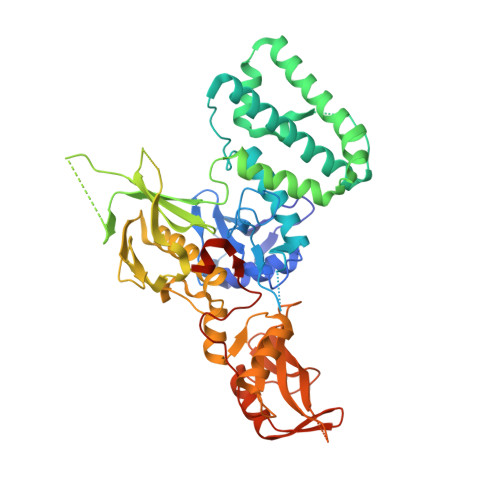
Crystal Structure of a Complex of the Intracellular Domain of Interferon lambda Receptor 1 (IFNLR1) and the FERM/SH2 Domains of Human JAK1.
Zhang, D., Wlodawer, A., Lubkowski, J.(2016) J Mol Biology 428: 4651-4668
- PubMed: 27725180
- DOI: https://doi.org/10.1016/j.jmb.2016.10.005
- Primary Citation of Related Structures:
5L04 - PubMed Abstract:
The crystal structure of a construct consisting of the FERM and SH2-like domains of the human Janus kinase 1 (JAK1) bound to a fragment of the intracellular domain of the interferon-λ receptor 1 (IFNLR1) has been determined at the nominal resolution of 2.1Å. In this structure, the receptor peptide forms an 85-Å-long extended chain, in which both the previously identified box1 and box2 regions bind simultaneously to the FERM and SH2-like domains of JAK1. Both domains of JAK1 are generally well ordered, with regions not seen in the crystal structure limited to loops located away from the receptor-binding regions. The structure provides a much more complete and accurate picture of the interactions between JAK1 and IFNLR1 than those given in earlier reports, illuminating the molecular basis of the JAK-cytokine receptor association. A glutamate residue adjacent to the box2 region in IFNLR1 mimics the mode of binding of a phosphotyrosine in classical SH2 domains. It was shown here that a deletion of residues within the box1 region of the receptor abolishes stable interactions with JAK1, although it was previously shown that box2 alone is sufficient to stabilize a similar complex of the interferon-α receptor and TYK2.
- Macromolecular Crystallography Laboratory, Center for Cancer Research, National Cancer Institute, Frederick, MD 21702, USA.
Organizational Affiliation: