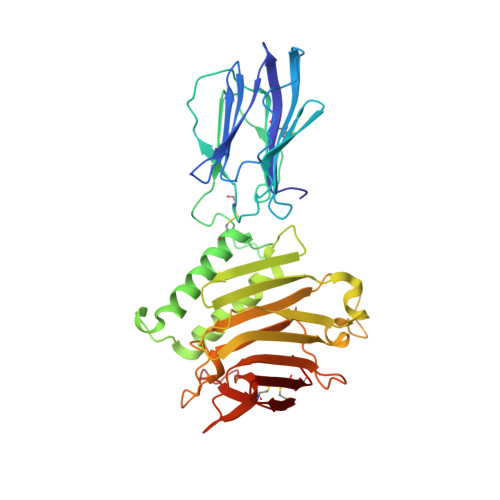
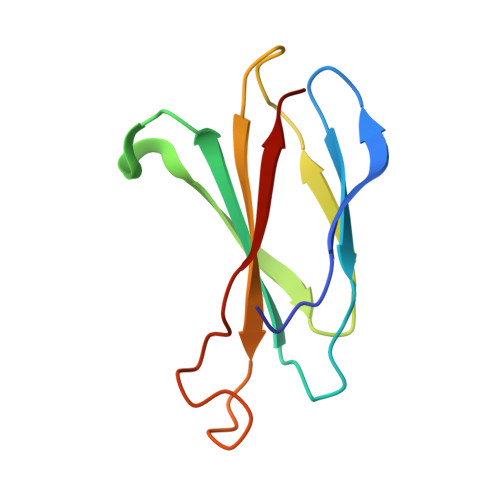
Structural Basis for Regulation of GPR56/ADGRG1 by Its Alternatively Spliced Extracellular Domains.
Salzman, G.S., Ackerman, S.D., Ding, C., Koide, A., Leon, K., Luo, R., Stoveken, H.M., Fernandez, C.G., Tall, G.G., Piao, X., Monk, K.R., Koide, S., Arac, D.(2016) Neuron 91: 1292-1304
- PubMed: 27657451
- DOI: https://doi.org/10.1016/j.neuron.2016.08.022
- Primary Citation of Related Structures:
5KVM - PubMed Abstract:
Adhesion G protein-coupled receptors (aGPCRs) play critical roles in diverse neurobiological processes including brain development, synaptogenesis, and myelination. aGPCRs have large alternatively spliced extracellular regions (ECRs) that likely mediate intercellular signaling; however, the precise roles of ECRs remain unclear. The aGPCR GPR56/ADGRG1 regulates both oligodendrocyte and cortical development. Accordingly, human GPR56 mutations cause myelination defects and brain malformations. Here, we determined the crystal structure of the GPR56 ECR, the first structure of any complete aGPCR ECR, in complex with an inverse-agonist monobody, revealing a GPCR-Autoproteolysis-Inducing domain and a previously unidentified domain that we term Pentraxin/Laminin/neurexin/sex-hormone-binding-globulin-Like (PLL). Strikingly, PLL domain deletion caused increased signaling and characterizes a GPR56 splice variant. Finally, we show that an evolutionarily conserved residue in the PLL domain is critical for oligodendrocyte development in vivo. Thus, our results suggest that the GPR56 ECR has unique and multifaceted regulatory functions, providing novel insights into aGPCR roles in neurobiology.
- Biophysical Sciences Program, The University of Chicago, Chicago, IL 60637, USA; Medical Scientist Training Program, The University of Chicago, Chicago, IL 60637, USA; Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL 60637, USA.
Organizational Affiliation: