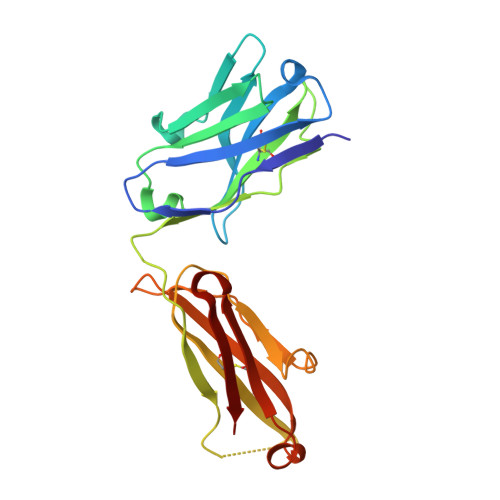
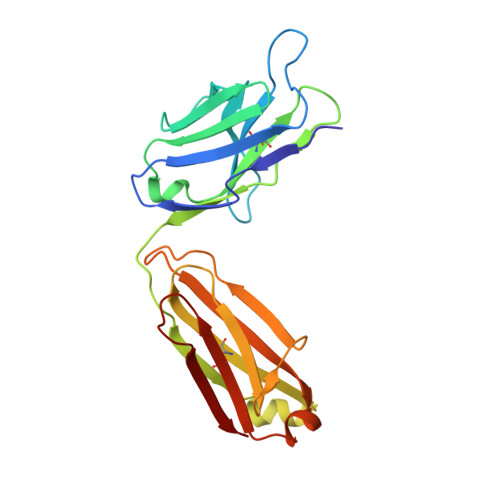
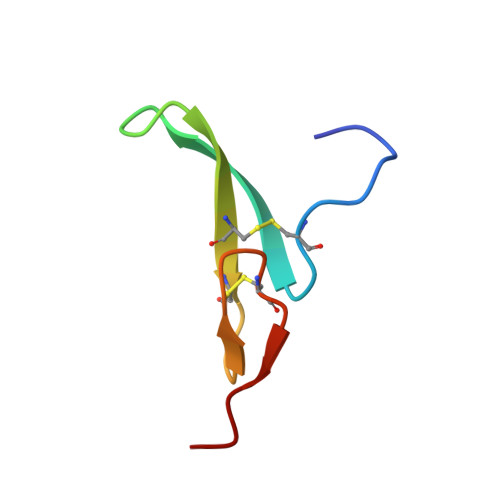
Structural basis of selectivity and neutralizing activity of a TGF alpha /epiregulin specific antibody.
Boyles, J.S., Atwell, S., Druzina, Z., Heuer, J.G., Witcher, D.R.(2016) Protein Sci 25: 2028-2036
- PubMed: 27543934
- DOI: https://doi.org/10.1002/pro.3023
- Primary Citation of Related Structures:
5KN5 - PubMed Abstract:
Recent studies have implicated a role of the epidermal growth factor receptor (EGFR) pathway in kidney disease. Skin toxicity associated with therapeutics which completely block the EGFR pathway precludes their use in chronic dosing. Therefore, we developed antibodies which specifically neutralize the EGFR ligands TGFα (transforming growth factor-alpha) and epiregulin but not EGF (epidermal growth factor), amphiregulin, betacellulin, HB-EGF (heparin-binding epidermal growth factor), or epigen. The epitope of one such neutralizing antibody, LY3016859, was characterized in detail to elucidate the structural basis for ligand specificity. Here we report a crystal structure of the LY3016859 Fab fragment in complex with soluble human TGFα. Our data demonstrate a conformational epitope located primarily within the C-terminal subdomain of the ligand. In addition, point mutagenesis experiments were used to highlight specific amino acids which are critical for both antigen binding and neutralization, most notably Ala 41 , Glu 44 , and His 45 . These results illustrate the structural basis for the ligand specificity/selectivity of LY3016859 and could also provide insight into further engineering to alter specificity and/or affinity of LY3016859.
- Biotechnology Discovery Research, Lilly Research Laboratories, Eli Lilly and Company, Indianapolis, Indiana, 46285.
Organizational Affiliation: