Discovery of 3-(3-(4-(1-Aminocyclobutyl)phenyl)-5-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amine (ARQ 092): An Orally Bioavailable, Selective, and Potent Allosteric AKT Inhibitor.
Lapierre, J.M., Eathiraj, S., Vensel, D., Liu, Y., Bull, C.O., Cornell-Kennon, S., Iimura, S., Kelleher, E.W., Kizer, D.E., Koerner, S., Makhija, S., Matsuda, A., Moussa, M., Namdev, N., Savage, R.E., Szwaya, J., Volckova, E., Westlund, N., Wu, H., Schwartz, B.(2016) J Med Chem 59: 6455-6469
- PubMed: 27305487
- DOI: https://doi.org/10.1021/acs.jmedchem.6b00619
- Primary Citation of Related Structures:
5KCV - PubMed Abstract:
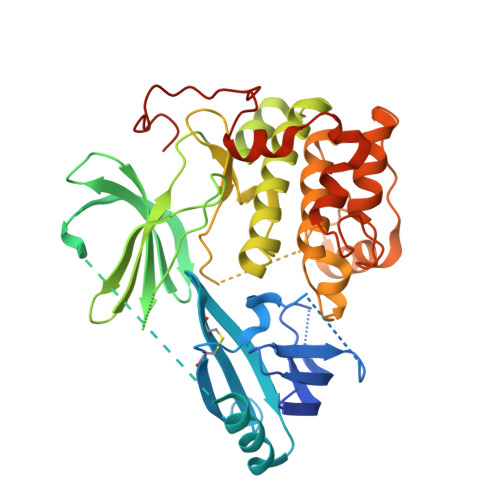
The work in this paper describes the optimization of the 3-(3-phenyl-3H-imidazo[4,5-b]pyridin-2-yl)pyridin-2-amine chemical series as potent, selective allosteric inhibitors of AKT kinases, leading to the discovery of ARQ 092 (21a). The cocrystal structure of compound 21a bound to full-length AKT1 confirmed the allosteric mode of inhibition of this chemical class and the role of the cyclobutylamine moiety. Compound 21a demonstrated high enzymatic potency against AKT1, AKT2, and AKT3, as well as potent cellular inhibition of AKT activation and the phosphorylation of the downstream target PRAS40. Compound 21a also served as a potent inhibitor of the AKT1-E17K mutant protein and inhibited tumor growth in a human xenograft mouse model of endometrial adenocarcinoma.
- ArQule Inc. , One Wall Street, Burlington, Massachusetts 01803, United States.
Organizational Affiliation: