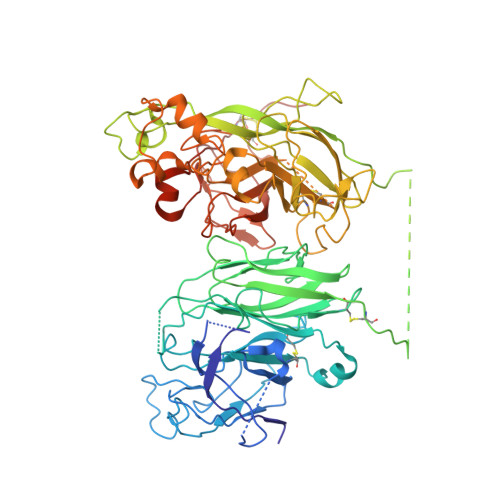
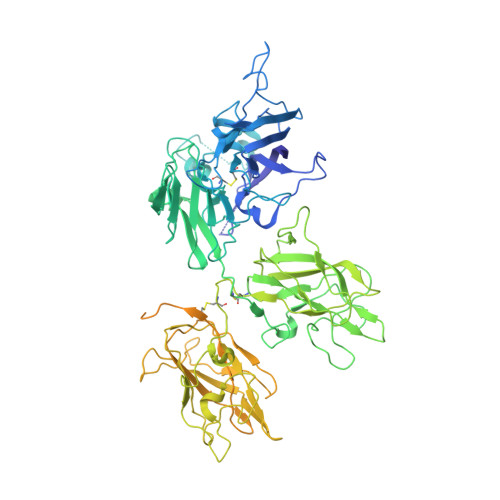
The structural basis for the functional comparability of factor VIII and the long-acting variant recombinant factor VIII Fc fusion protein.
Leksa, N.C., Chiu, P.L., Bou-Assaf, G.M., Quan, C., Liu, Z., Goodman, A.B., Chambers, M.G., Tsutakawa, S.E., Hammel, M., Peters, R.T., Walz, T., Kulman, J.D.(2017) J Thromb Haemost 15: 1167-1179
- PubMed: 28397397
- DOI: https://doi.org/10.1111/jth.13700
- Primary Citation of Related Structures:
5K8D - PubMed Abstract:
Essentials Recombinant factor VIII (rFVIII) Fc fusion protein has a 1.5-fold longer half-life than rFVIII. Five orthogonal methods were used to characterize the structure of rFVIIIFc compared to rFVIII. The C-terminal Fc fusion does not perturb the structure of FVIII in rFVIIIFc. The FVIII and Fc components of rFVIIIFc are flexibly tethered and functionally independent. Background Fusion of the human IgG 1 Fc domain to the C-terminal C2 domain of B-domain-deleted (BDD) factor VIII (FVIII) results in the recombinant FVIII Fc (rFVIIIFc) fusion protein, which has a 1.5-fold longer half-life in humans. Objective To assess the structural properties of rFVIIIFc by comparing its constituent FVIII and Fc elements with their respective isolated components, and evaluating their structural independence within rFVIIIFc. Methods rFVIIIFc and its isolated FVIII and Fc components were compared by the use of hydrogen-deuterium exchange mass spectrometry (HDX-MS). The structure of rFVIIIFc was also evaluated by the use of X-ray crystallography, small-angle X-ray scattering (SAXS), and electron microscopy (EM). The degree of steric interference by the appended Fc domain was assessed by EM and surface plasmon resonance (SPR). Results HDX-MS analysis of rFVIIIFc revealed that fusion caused no structural perturbations in FVIII or Fc. The rFVIIIFc crystal structure showed that the FVIII component is indistinguishable from published BDD FVIII structures. The Fc domain was not observed, indicating high mobility. SAXS analysis was consistent with an ensemble of rigid-body models in which the Fc domain exists in a largely extended orientation relative to FVIII. Binding of Fab fragments of anti-C2 domain antibodies to BDD FVIII was visualized by EM, and the affinities of the corresponding intact antibodies for BDD FVIII and rFVIIIFc were comparable by SPR analysis. Conclusions The FVIII and Fc components of rFVIIIFc are structurally indistinguishable from their isolated constituents, and show a high degree of structural independence, consistent with the functional comparability of rFVIIIFc and unmodified FVIII.
- Biogen, Cambridge, MA, USA.
Organizational Affiliation: