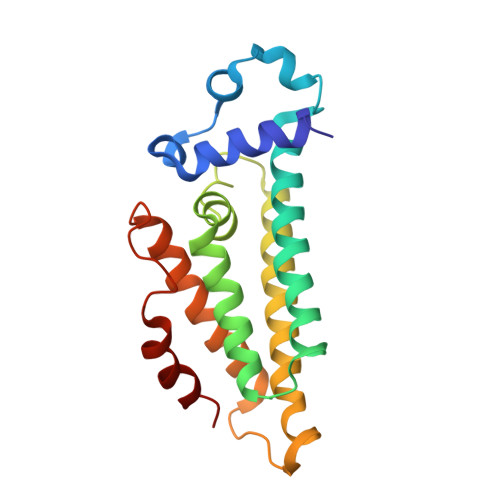
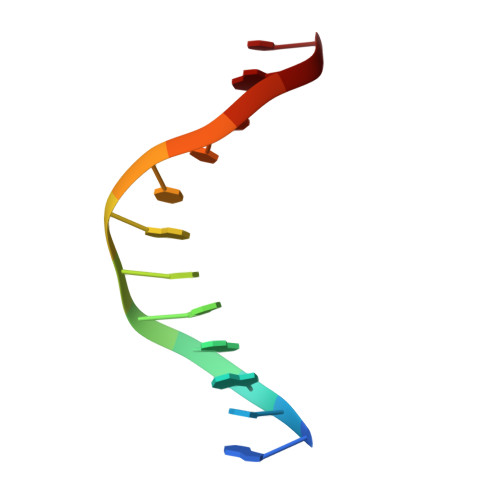
Structures of the nucleoid occlusion protein SlmA bound to DNA and the C-terminal domain of the cytoskeletal protein FtsZ.
Schumacher, M.A., Zeng, W.(2016) Proc Natl Acad Sci U S A 113: 4988-4993
- PubMed: 27091999
- DOI: https://doi.org/10.1073/pnas.1602327113
- Primary Citation of Related Structures:
5HAW, 5HBU, 5HSZ, 5K58 - PubMed Abstract:
Cell division in most prokaryotes is mediated by FtsZ, which polymerizes to create the cytokinetic Z ring. Multiple FtsZ-binding proteins regulate FtsZ polymerization to ensure the proper spatiotemporal formation of the Z ring at the division site. The DNA-binding protein SlmA binds to FtsZ and prevents Z-ring formation through the nucleoid in a process called "nucleoid occlusion" (NO). As do most FtsZ-accessory proteins, SlmA interacts with the conserved C-terminal domain (CTD) that is connected to the FtsZ core by a long, flexible linker. However, SlmA is distinct from other regulatory factors in that it must be DNA-bound to interact with the FtsZ CTD. Few structures of FtsZ regulator-CTD complexes are available, but all reveal the CTD bound as a helix. To deduce the molecular basis for the unique SlmA-DNA-FtsZ CTD regulatory interaction and provide insight into FtsZ-regulator protein complex formation, we determined structures of Escherichia coli, Vibrio cholera, and Klebsiella pneumonia SlmA-DNA-FtsZ CTD ternary complexes. Strikingly, the FtsZ CTD does not interact with SlmA as a helix but binds as an extended conformation in a narrow, surface-exposed pocket formed only in the DNA-bound state of SlmA and located at the junction between the DNA-binding and C-terminal dimer domains. Binding studies are consistent with the structure and underscore key interactions in complex formation. Combined, these data reveal the molecular basis for the SlmA-DNA-FtsZ interaction with implications for SlmA's NO function and underscore the ability of the FtsZ CTD to adopt a wide range of conformations, explaining its ability to bind diverse regulatory proteins.
- Department of Biochemistry, Duke University School of Medicine, Durham, NC 27710 maria.schumacher@duke.edu.
Organizational Affiliation: