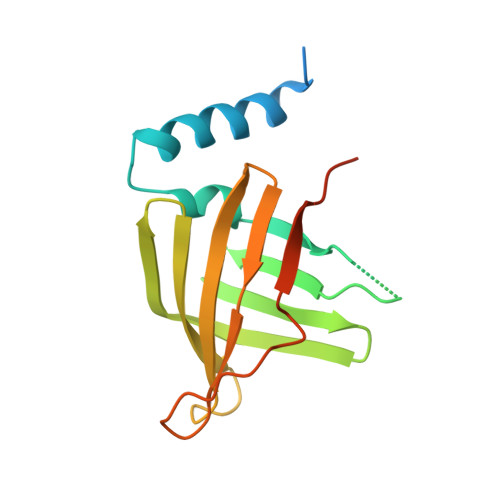
The Structure of Treponema pallidum Tp0751 (Pallilysin) Reveals a Non-canonical Lipocalin Fold That Mediates Adhesion to Extracellular Matrix Components and Interactions with Host Cells.
Parker, M.L., Houston, S., Petrosova, H., Lithgow, K.V., Hof, R., Wetherell, C., Kao, W.C., Lin, Y.P., Moriarty, T.J., Ebady, R., Cameron, C.E., Boulanger, M.J.(2016) PLoS Pathog 12: e1005919-e1005919
- PubMed: 27683203
- DOI: https://doi.org/10.1371/journal.ppat.1005919
- Primary Citation of Related Structures:
5JK2 - PubMed Abstract:
Syphilis is a chronic disease caused by the bacterium Treponema pallidum subsp. pallidum. Treponema pallidum disseminates widely throughout the host and extravasates from the vasculature, a process that is at least partially dependent upon the ability of T. pallidum to interact with host extracellular matrix (ECM) components. Defining the molecular basis for the interaction between T. pallidum and the host is complicated by the intractability of T. pallidum to in vitro culturing and genetic manipulation. Correspondingly, few T. pallidum proteins have been identified that interact directly with host components. Of these, Tp0751 (also known as pallilysin) displays a propensity to interact with the ECM, although the underlying mechanism of these interactions remains unknown. Towards establishing the molecular mechanism of Tp0751-host ECM attachment, we first determined the crystal structure of Tp0751 to a resolution of 2.15 Å using selenomethionine phasing. Structural analysis revealed an eight-stranded beta-barrel with a profile of short conserved regions consistent with a non-canonical lipocalin fold. Using a library of native and scrambled peptides representing the full Tp0751 sequence, we next identified a subset of peptides that showed statistically significant and dose-dependent interactions with the ECM components fibrinogen, fibronectin, collagen I, and collagen IV. Intriguingly, each ECM-interacting peptide mapped to the lipocalin domain. To assess the potential of these ECM-coordinating peptides to inhibit adhesion of bacteria to host cells, we engineered an adherence-deficient strain of the spirochete Borrelia burgdorferi to heterologously express Tp0751. This engineered strain displayed Tp0751 on its surface and exhibited a Tp0751-dependent gain-of-function in adhering to human umbilical vein endothelial cells that was inhibited in the presence of one of the ECM-interacting peptides (p10). Overall, these data provide the first structural insight into the mechanisms of Tp0751-host interactions, which are dependent on the protein's lipocalin fold.
- Department of Biochemistry and Microbiology, University of Victoria, Victoria, British Columbia, Canada.
Organizational Affiliation: