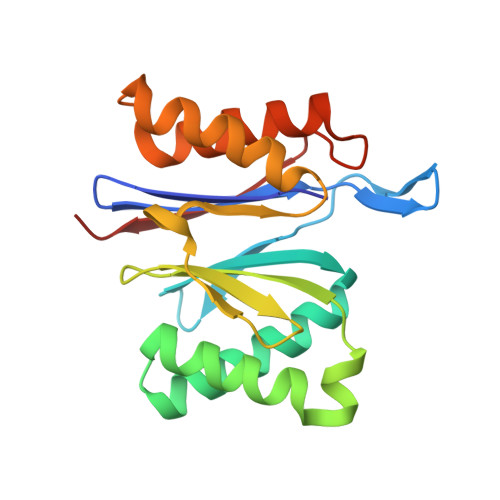
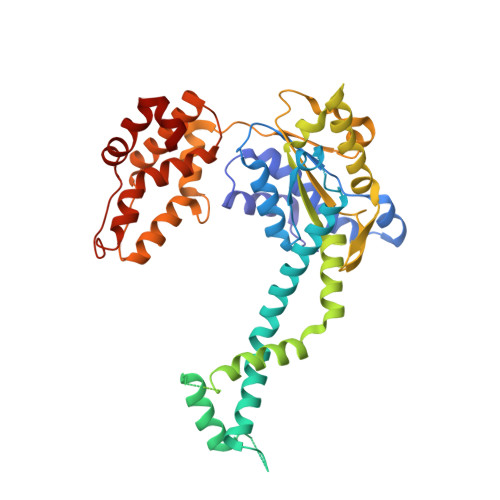
A Structurally Dynamic Region of the HslU Intermediate Domain Controls Protein Degradation and ATP Hydrolysis.
Baytshtok, V., Fei, X., Grant, R.A., Baker, T.A., Sauer, R.T.(2016) Structure 24: 1766-1777
- PubMed: 27667691
- DOI: https://doi.org/10.1016/j.str.2016.08.012
- Primary Citation of Related Structures:
5JI2, 5JI3 - PubMed Abstract:
The I domain of HslU sits above the AAA+ ring and forms a funnel-like entry to the axial pore, where protein substrates are engaged, unfolded, and translocated into HslV for degradation. The L199Q I-domain substitution, which was originally reported as a loss-of-function mutation, resides in a segment that appears to adopt multiple conformations as electron density is not observed in HslU and HslUV crystal structures. The L199Q sequence change does not alter the structure of the AAA+ ring or its interactions with HslV but increases I-domain susceptibility to limited endoproteolysis. Notably, the L199Q mutation increases the rate of ATP hydrolysis substantially, results in slower degradation of some proteins but faster degradation of other substrates, and markedly changes the preference of HslUV for initiating degradation at the N or C terminus of model substrates. Thus, a structurally dynamic region of the I domain plays a key role in controlling protein degradation by HslUV.
- Department of Biology, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
Organizational Affiliation: