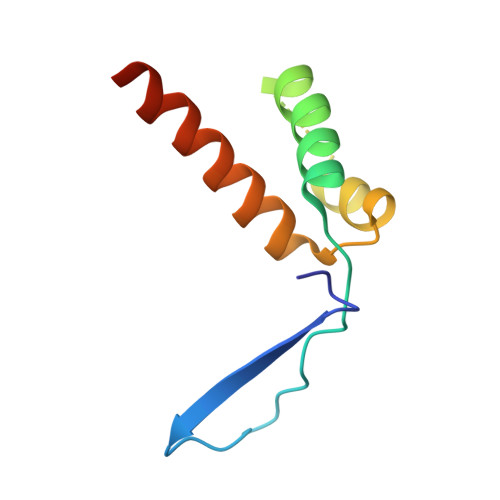
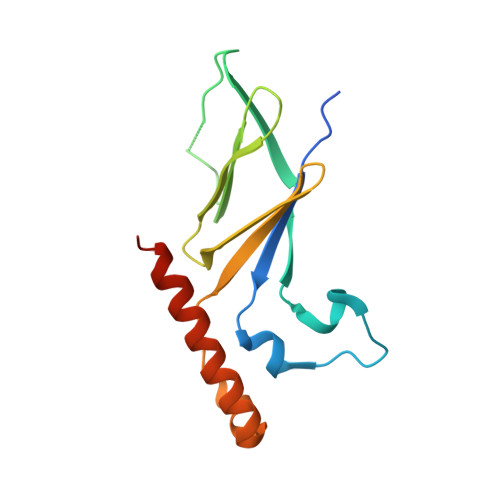
The Intrinsically Disordered Protein Atg13 Mediates Supramolecular Assembly of Autophagy Initiation Complexes.
Yamamoto, H., Fujioka, Y., Suzuki, S.W., Noshiro, D., Suzuki, H., Kondo-Kakuta, C., Kimura, Y., Hirano, H., Ando, T., Noda, N.N., Ohsumi, Y.(2016) Dev Cell 38: 86-99
- PubMed: 27404361
- DOI: https://doi.org/10.1016/j.devcel.2016.06.015
- Primary Citation of Related Structures:
5JHF - PubMed Abstract:
Autophagosome formation in yeast entails starvation-induced assembly of the pre-autophagosomal structure (PAS), in which multiple Atg1 complexes (composed of Atg1, Atg13, and the Atg17-Atg29-Atg31 subcomplex) are initially engaged. However, the molecular mechanisms underlying the multimeric assembly of these complexes remain unclear. Using structural and biological techniques, we herein demonstrate that Atg13 has a large intrinsically disordered region (IDR) and interacts with two distinct Atg17 molecules using two binding regions in the IDR. We further reveal that these two binding regions are essential not only for Atg1 complex assembly in vitro, but also for PAS organization in vivo. These findings underscore the structural and functional significance of the IDR of Atg13 in autophagy initiation: Atg13 provides intercomplex linkages between Atg17-Atg29-Atg31 complexes, thereby leading to supramolecular self-assembly of Atg1 complexes, in turn accelerating the initial events of autophagy, including autophosphorylation of Atg1, recruitment of Atg9 vesicles, and phosphorylation of Atg9 by Atg1.
- Frontier Research Center, Tokyo Institute of Technology, Yokohama 226-8503, Japan.
Organizational Affiliation: