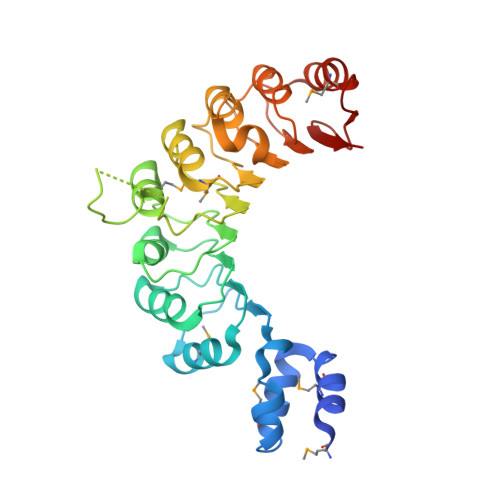
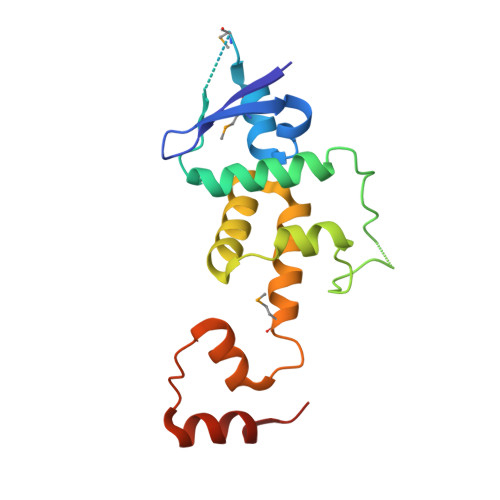
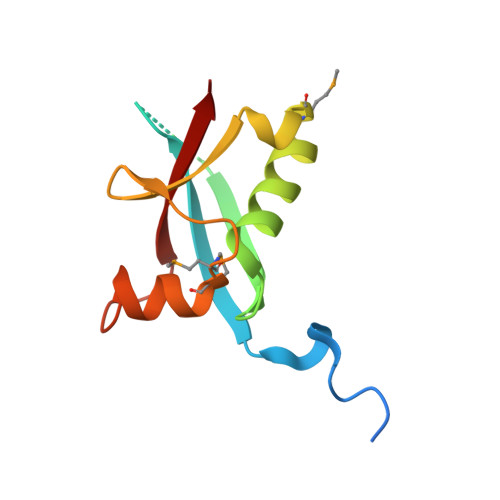
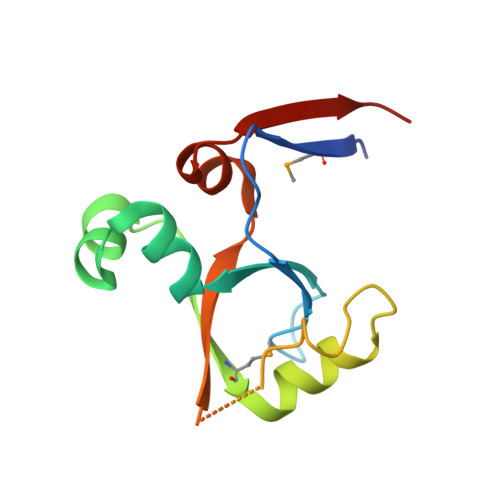
KDM2B Recruitment of the Polycomb Group Complex, PRC1.1, Requires Cooperation between PCGF1 and BCORL1.
Wong, S.J., Gearhart, M.D., Taylor, A.B., Nanyes, D.R., Ha, D.J., Robinson, A.K., Artigas, J.A., Lee, O.J., Demeler, B., Hart, P.J., Bardwell, V.J., Kim, C.A.(2016) Structure 24: 1795-1801
- PubMed: 27568929
- DOI: https://doi.org/10.1016/j.str.2016.07.011
- Primary Citation of Related Structures:
5JH5 - PubMed Abstract:
KDM2B recruits H2A-ubiquitinating activity of a non-canonical Polycomb Repression Complex 1 (PRC1.1) to CpG islands, facilitating gene repression. We investigated the molecular basis of recruitment using in vitro assembly assays to identify minimal components, subcomplexes, and domains required for recruitment. A minimal four-component PRC1.1 complex can be assembled by combining two separately isolated subcomplexes: the DNA-binding KDM2B/SKP1 heterodimer and the heterodimer of BCORL1 and PCGF1, a core component of PRC1.1. The crystal structure of the KDM2B/SKP1/BCORL1/PCGF1 complex illustrates the crucial role played by the PCGF1/BCORL1 heterodimer. The BCORL1 PUFD domain positions residues preceding the RAWUL domain of PCGF1 to create an extended interface for interaction with KDM2B, which is unique to the PCGF1-containing PRC1.1 complex. The structure also suggests how KDM2B might simultaneously function in PRC1.1 and an SCF ubiquitin ligase complex and the possible molecular consequences of BCOR PUFD internal tandem duplications found in pediatric kidney and brain tumors.
- Department of Biochemistry and CTRC, University of Texas Health Science Center at San Antonio, MSC 7760, 7703 Floyd Curl Drive, San Antonio, TX 78229-3990, USA.
Organizational Affiliation: