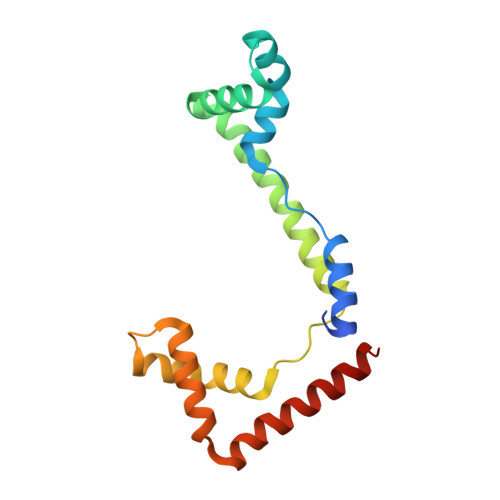
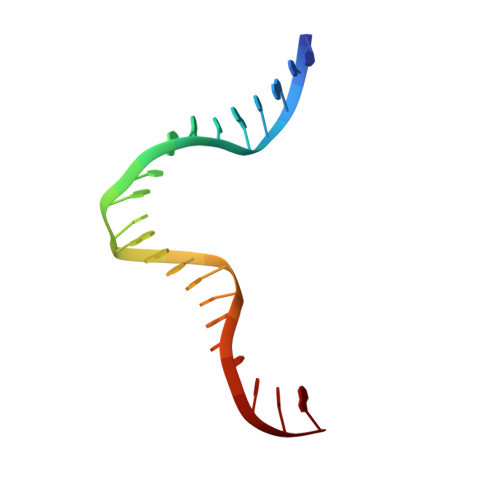
DNA structure directs positioning of the mitochondrial genome packaging protein Abf2p.
Chakraborty, A., Lyonnais, S., Battistini, F., Hospital, A., Medici, G., Prohens, R., Orozco, M., Vilardell, J., Sola, M.(2017) Nucleic Acids Res 45: 951-967
- PubMed: 27899643
- DOI: https://doi.org/10.1093/nar/gkw1147
- Primary Citation of Related Structures:
5JGH, 5JH0 - PubMed Abstract:
The mitochondrial genome (mtDNA) is assembled into nucleo-protein structures termed nucleoids and maintained differently compared to nuclear DNA, the involved molecular basis remaining poorly understood. In yeast (Saccharomyces cerevisiae), mtDNA is a ∼80 kbp linear molecule and Abf2p, a double HMG-box protein, packages and maintains it. The protein binds DNA in a non-sequence-specific manner, but displays a distinct 'phased-binding' at specific DNA sequences containing poly-adenine tracts (A-tracts). We present here two crystal structures of Abf2p in complex with mtDNA-derived fragments bearing A-tracts. Each HMG-box of Abf2p induces a 90° bend in the contacted DNA, causing an overall U-turn. Together with previous data, this suggests that U-turn formation is the universal mechanism underlying mtDNA compaction induced by HMG-box proteins. Combining this structural information with mutational, biophysical and computational analyses, we reveal a unique DNA binding mechanism for Abf2p where a characteristic N-terminal flag and helix are crucial for mtDNA maintenance. Additionally, we provide the molecular basis for A-tract mediated exclusion of Abf2p binding. Due to high prevalence of A-tracts in yeast mtDNA, this has critical relevance for nucleoid architecture. Therefore, an unprecedented A-tract mediated protein positioning mechanism regulates DNA packaging proteins in the mitochondria, and in combination with DNA-bending and U-turn formation, governs mtDNA compaction.
- Structural MitoLab, Department of Structural Biology, "Maria de Maeztu" Unit of Excellence, Molecular Biology Institute Barcelona (IBMB-CSIC), Barcelona 08028, Spain.
Organizational Affiliation: