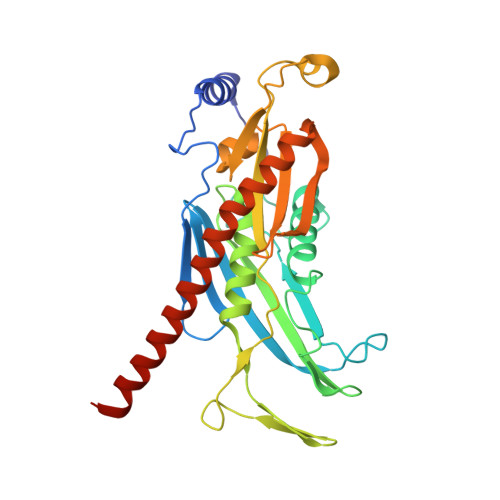
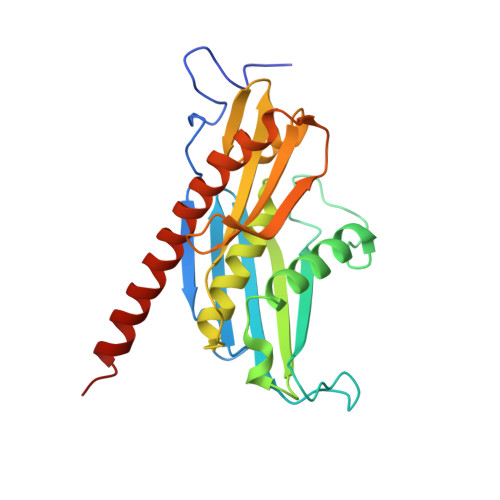
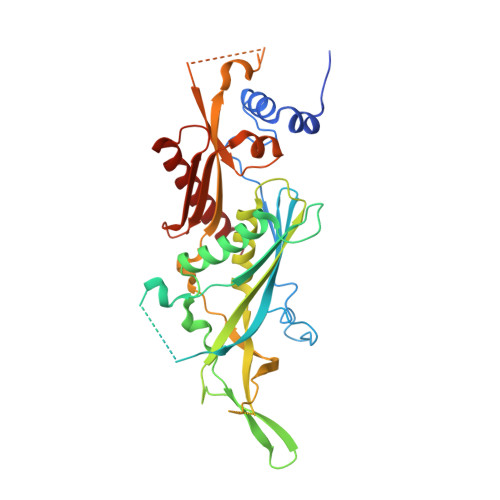
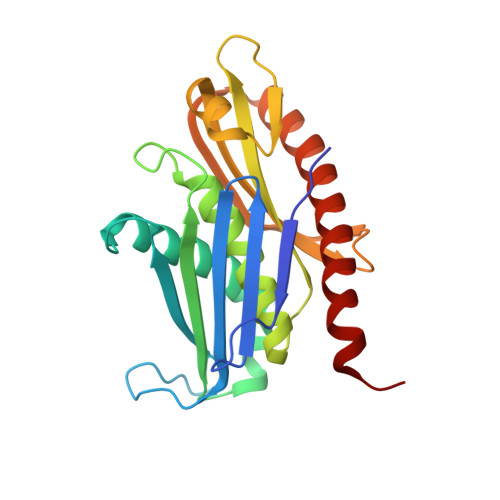
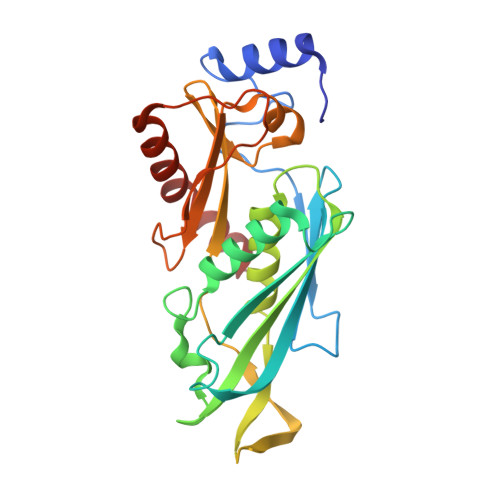
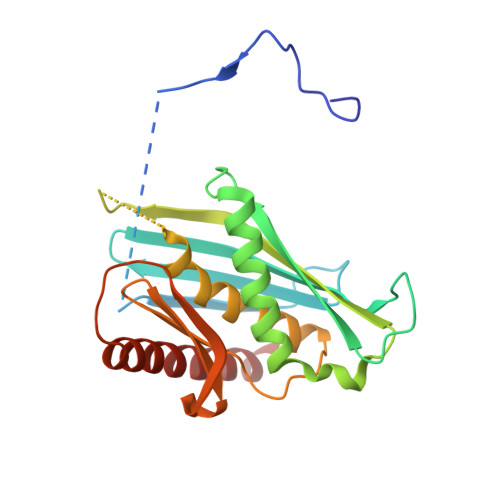
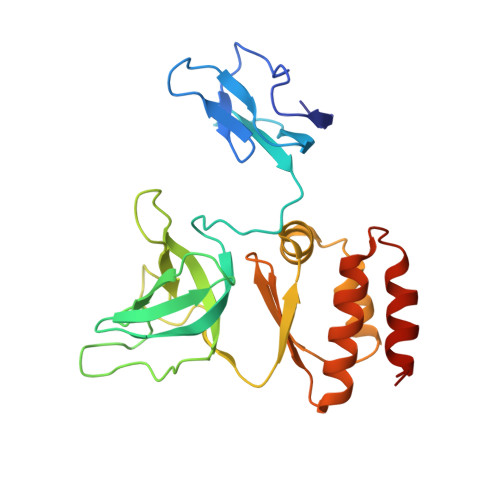
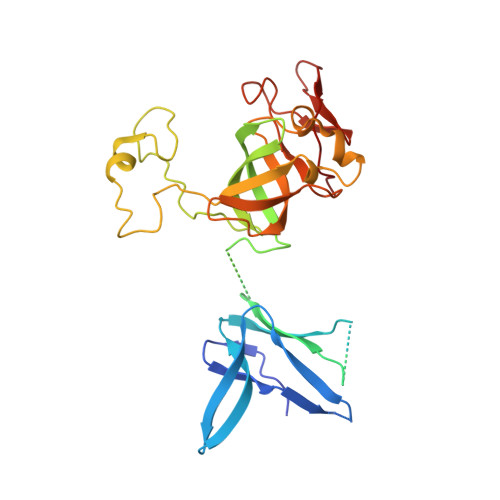
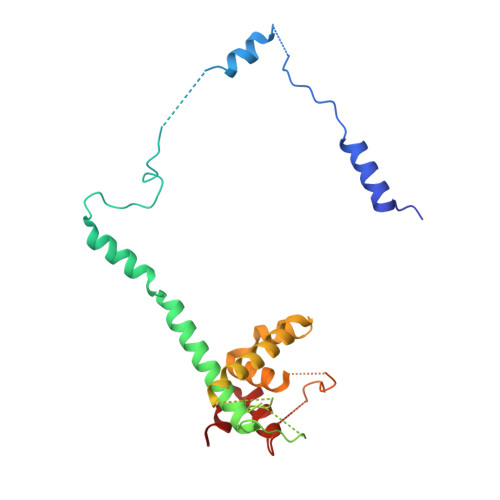
Structure of a Cytoplasmic 11-Subunit RNA Exosome Complex.
Kowalinski, E., Kogel, A., Ebert, J., Reichelt, P., Stegmann, E., Habermann, B., Conti, E.(2016) Mol Cell 63: 125-134
- PubMed: 27345150
- DOI: https://doi.org/10.1016/j.molcel.2016.05.028
- Primary Citation of Related Structures:
5JEA - PubMed Abstract:
The RNA exosome complex associates with nuclear and cytoplasmic cofactors to mediate the decay, surveillance, or processing of a wide variety of transcripts. In the cytoplasm, the conserved core of the exosome (Exo10) functions together with the conserved Ski complex. The interaction of S. cerevisiae Exo10 and Ski is not direct but requires a bridging cofactor, Ski7. Here, we report the 2.65 Å resolution structure of S. cerevisiae Exo10 bound to the interacting domain of Ski7. Extensive hydrophobic interactions rationalize the high affinity and stability of this complex, pointing to Ski7 as a constitutive component of the cytosolic exosome. Despite the absence of sequence homology, cytoplasmic Ski7 and nuclear Rrp6 bind Exo10 using similar surfaces and recognition motifs. Knowledge of the interacting residues in the yeast complexes allowed us to identify a splice variant of human HBS1-Like as a Ski7-like exosome-binding protein, revealing the evolutionary conservation of this cytoplasmic cofactor.
- Department of Structural Cell Biology, Max Planck Institute of Biochemistry, Am Klopferspitz 18, 82152 Martinsried, Germany.
Organizational Affiliation: