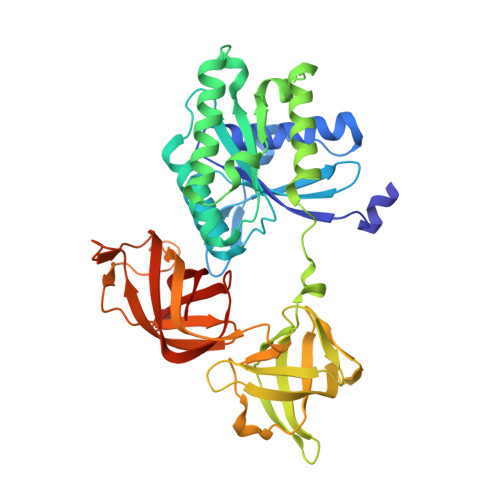
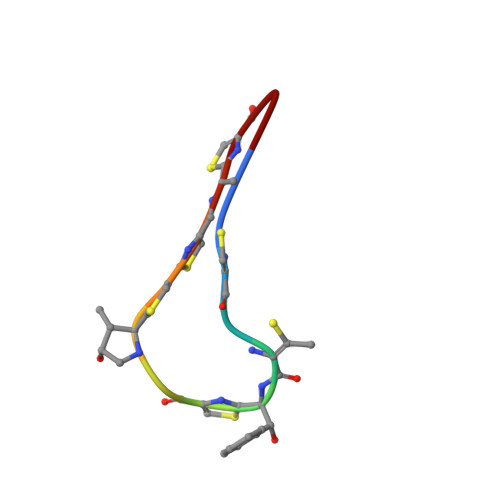
Antibacterial and Solubility Optimization of Thiomuracin A.
LaMarche, M.J., Leeds, J.A., Brewer, J., Dean, K., Ding, J., Dzink-Fox, J., Gamber, G., Jain, A., Kerrigan, R., Krastel, P., Lee, K., Lombardo, F., McKenney, D., Neckermann, G., Osborne, C., Palestrant, D., Patane, M.A., Rann, E.M., Robinson, Z., Schmitt, E., Stams, T., Tiamfook, S., Yu, D., Whitehead, L.(2016) J Med Chem 59: 6920-6928
- PubMed: 27355833
- DOI: https://doi.org/10.1021/acs.jmedchem.6b00726
- Primary Citation of Related Structures:
5JBQ - PubMed Abstract:
Synthetic studies of the antimicrobial secondary metabolite thiomuracin A (1) provided access to analogues in the Northern region (C2-C10). Selective hydrolysis of the C10 amide of lead compound 2 and subsequent derivatization led to novel carbon- and nitrogen-linked analogues (e.g., 3) which improved antibacterial potency across a panel of Gram-positive organisms. In addition, congeners with improved physicochemical properties were identified which proved efficacious in murine sepsis and hamster C. difficile models of disease. Optimal efficacy in the hamster model of C. difficile was achieved with compounds that possessed both potent antibacterial activity and high aqueous solubility.
- Infectious Disease Area, Novartis Institutes for Biomedical Research , Emeryville, California 94608, United States.
Organizational Affiliation: