Recognition of the Major Histocompatibility Complex (MHC) Class Ib Molecule H2-Q10 by the Natural Killer Cell Receptor Ly49C.
Sullivan, L.C., Berry, R., Sosnin, N., Widjaja, J.M., Deuss, F.A., Balaji, G.R., LaGruta, N.L., Mirams, M., Trapani, J.A., Rossjohn, J., Brooks, A.G., Andrews, D.M.(2016) J Biological Chem 291: 18740-18752
- PubMed: 27385590
- DOI: https://doi.org/10.1074/jbc.M116.737130
- Primary Citation of Related Structures:
5J6G, 5J6H - PubMed Abstract:
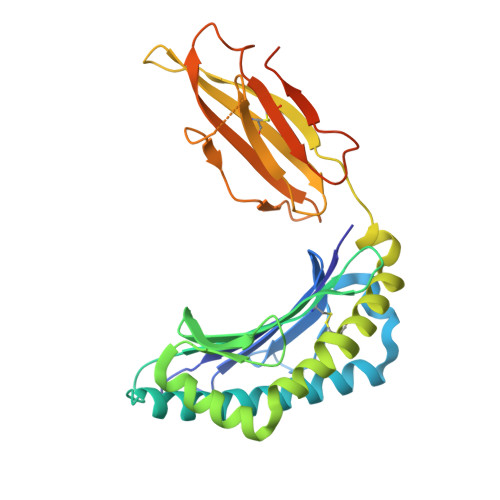
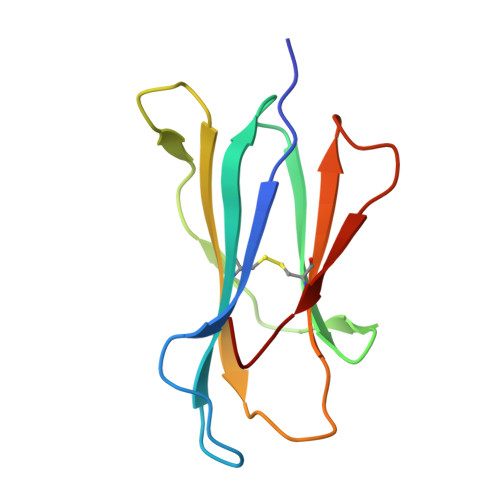
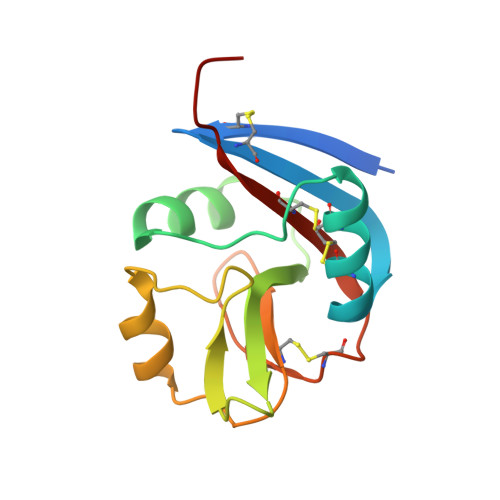
Murine natural killer (NK) cells are regulated by the interaction of Ly49 receptors with major histocompatibility complex class I molecules (MHC-I). Although the ligands for inhibitory Ly49 were considered to be restricted to classical MHC (MHC-Ia), we have shown that the non-classical MHC molecule (MHC-Ib) H2-M3 was a ligand for the inhibitory Ly49A. Here we establish that another MHC-Ib, H2-Q10, is a bona fide ligand for the inhibitory Ly49C receptor. H2-Q10 bound to Ly49C with a marginally lower affinity (∼5 μm) than that observed between Ly49C and MHC-Ia (H-2K(b)/H-2D(d), both ∼1 μm), and this recognition could be prevented by cis interactions with H-2K in situ To understand the molecular details underpinning Ly49·MHC-Ib recognition, we determined the crystal structures of H2-Q10 and Ly49C bound H2-Q10. Unliganded H2-Q10 adopted a classical MHC-I fold and possessed a peptide-binding groove that exhibited features similar to those found in MHC-Ia, explaining the diverse peptide binding repertoire of H2-Q10. Ly49C bound to H2-Q10 underneath the peptide binding platform to a region that encompassed residues from the α1, α2, and α3 domains, as well as the associated β2-microglobulin subunit. This docking mode was conserved with that previously observed for Ly49C·H-2K(b) Indeed, structure-guided mutation of Ly49C indicated that Ly49C·H2-Q10 and Ly49C·H-2K(b) possess similar energetic footprints focused around residues located within the Ly49C β4-stand and L5 loop, which contact the underside of the peptide-binding platform floor. Our data provide a structural basis for Ly49·MHC-Ib recognition and demonstrate that MHC-Ib represent an extended family of ligands for Ly49 molecules.
- From the Department of Microbiology and Immunology, University of Melbourne, Parkville, Victoria 3010, Australia.
Organizational Affiliation: