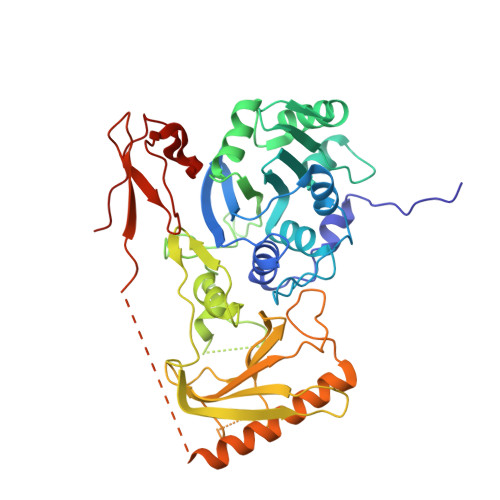
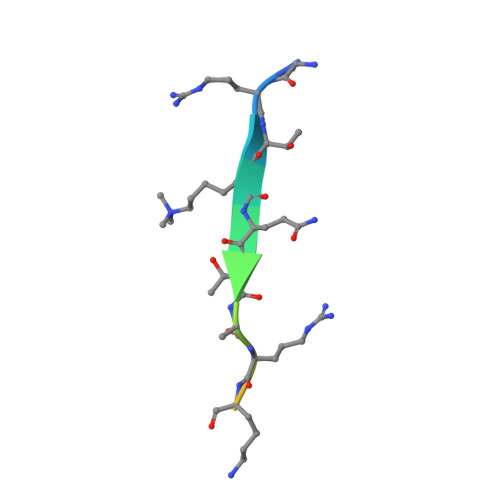
Mouse MORC3 is a GHKL ATPase that localizes to H3K4me3 marked chromatin
Li, S., Yen, L., Pastor, W.A., Johnston, J.B., Du, J., Shew, C.J., Liu, W., Ho, J., Stender, B., Clark, A.T., Burlingame, A.L., Daxinger, L., Patel, D.J., Jacobsen, S.E.(2016) Proc Natl Acad Sci U S A 113: E5108-E5116
- PubMed: 27528681
- DOI: https://doi.org/10.1073/pnas.1609709113
- Primary Citation of Related Structures:
5IX1, 5IX2 - PubMed Abstract:
Microrchidia (MORC) proteins are GHKL (gyrase, heat-shock protein 90, histidine kinase, MutL) ATPases that function in gene regulation in multiple organisms. Animal MORCs also contain CW-type zinc finger domains, which are known to bind to modified histones. We solved the crystal structure of the murine MORC3 ATPase-CW domain bound to the nucleotide analog AMPPNP (phosphoaminophosphonic acid-adenylate ester) and in complex with a trimethylated histone H3 lysine 4 (H3K4) peptide (H3K4me3). We observed that the MORC3 N-terminal ATPase domain forms a dimer when bound to AMPPNP. We used native mass spectrometry to show that dimerization is ATP-dependent, and that dimer formation is enhanced in the presence of nonhydrolyzable ATP analogs. The CW domain uses an aromatic cage to bind trimethylated Lys4 and forms extensive hydrogen bonds with the H3 tail. We found that MORC3 localizes to promoters marked by H3K4me3 throughout the genome, consistent with its binding to H3K4me3 in vitro. Our work sheds light on aspects of the molecular dynamics and function of MORC3.
- Structural Biology Program, Memorial Sloan Kettering Cancer Center, New York, NY 10065;
Organizational Affiliation: