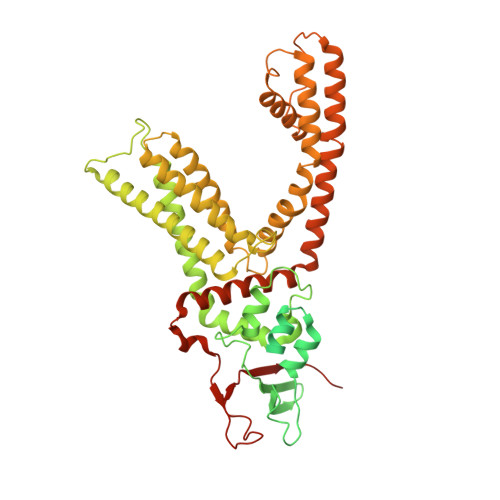
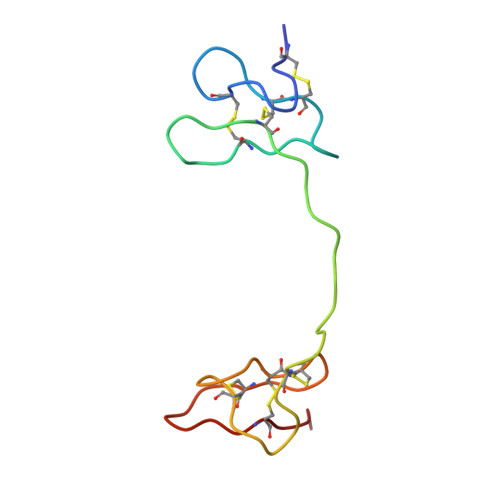
TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.
Gao, Y., Cao, E., Julius, D., Cheng, Y.(2016) Nature 534: 347-351
- PubMed: 27281200
- DOI: https://doi.org/10.1038/nature17964
- Primary Citation of Related Structures:
5IRX, 5IRZ, 5IS0 - PubMed Abstract:
When integral membrane proteins are visualized in detergents or other artificial systems, an important layer of information is lost regarding lipid interactions and their effects on protein structure. This is especially relevant to proteins for which lipids have both structural and regulatory roles. Here we demonstrate the power of combining electron cryo-microscopy with lipid nanodisc technology to ascertain the structure of the rat TRPV1 ion channel in a native bilayer environment. Using this approach, we determined the locations of annular and regulatory lipids and showed that specific phospholipid interactions enhance binding of a spider toxin to TRPV1 through formation of a tripartite complex. Furthermore, phosphatidylinositol lipids occupy the binding site for capsaicin and other vanilloid ligands, suggesting a mechanism whereby chemical or thermal stimuli elicit channel activation by promoting the release of bioactive lipids from a critical allosteric regulatory site.
- Department of Physiology, University of California, San Francisco, California 94143, USA.
Organizational Affiliation: