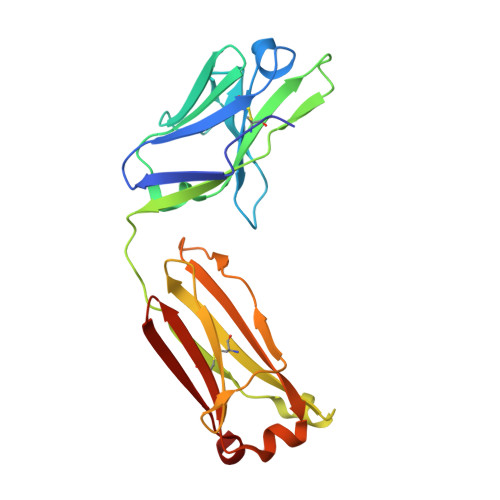
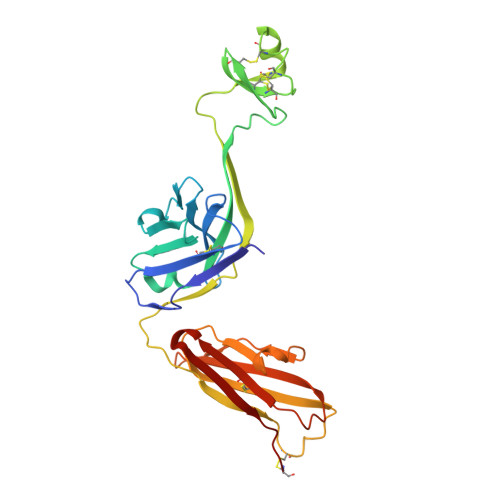
Conservation and diversity in the ultralong third heavy-chain complementarity-determining region of bovine antibodies.
Stanfield, R.L., Wilson, I.A., Smider, V.V.(2016) Sci Immunol 1
- PubMed: 27574710
- DOI: https://doi.org/10.1126/sciimmunol.aaf7962
- Primary Citation of Related Structures:
5IHU, 5IJV, 5ILT - PubMed Abstract:
A subset of bovine antibodies have an exceptionally long third heavy-chain complementarity determining region (CDR H3) that is highly variable in sequence and includes multiple cysteines. These long CDR H3s (up to 69 residues) fold into a long stalk atop which sits a knob domain that is located far from the antibody surface. Three new bovine Fab crystal structures have been determined to decipher the conserved and variable features of ultralong CDR H3s that lead to diversity in antigen recognition. Despite high sequence variability, the stalks adopt a conserved β-ribbon structure, while the knob regions share a conserved β-sheet that serves as a scaffold for two connecting loops of variable length and conformation, as well as one conserved disulfide. Variation in patterns and connectivity of the remaining disulfides contribute to the knob structural diversity. The unusual architecture of these ultralong bovine CDR H3s for generating diversity is unique in adaptive immune systems.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, California, 92037, USA.
Organizational Affiliation: