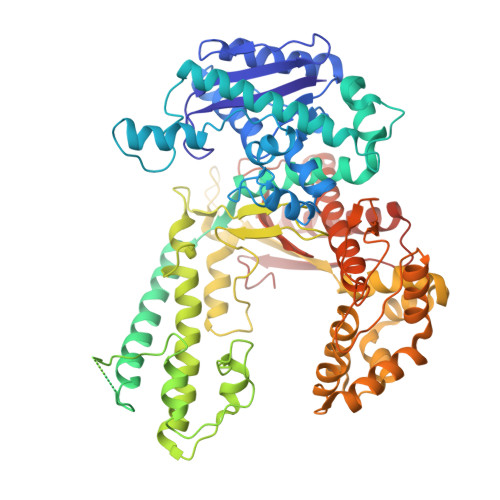
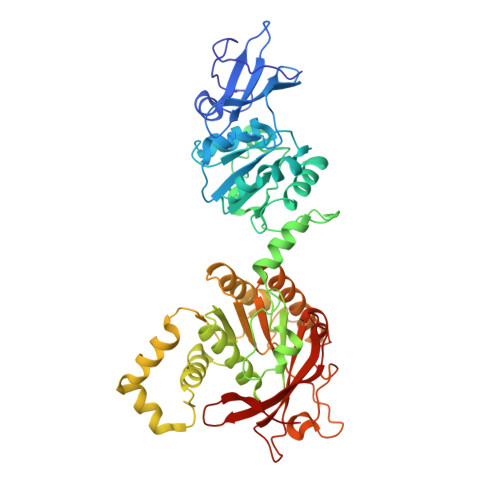
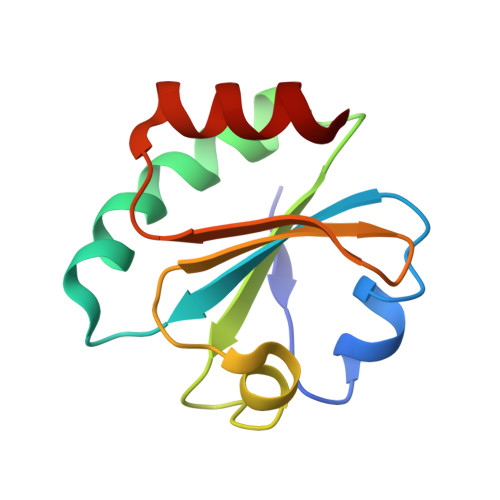
Hybrid Methods Reveal Multiple Flexibly Linked DNA Polymerases within the Bacteriophage T7 Replisome.
Wallen, J.R., Zhang, H., Weis, C., Cui, W., Foster, B.M., Ho, C.M., Hammel, M., Tainer, J.A., Gross, M.L., Ellenberger, T.(2017) Structure 25: 157-166
- PubMed: 28052235
- DOI: https://doi.org/10.1016/j.str.2016.11.019
- Primary Citation of Related Structures:
5IKN - PubMed Abstract:
The physical organization of DNA enzymes at a replication fork enables efficient copying of two antiparallel DNA strands, yet dynamic protein interactions within the replication complex complicate replisome structural studies. We employed a combination of crystallographic, native mass spectrometry and small-angle X-ray scattering experiments to capture alternative structures of a model replication system encoded by bacteriophage T7. Two molecules of DNA polymerase bind the ring-shaped primase-helicase in a conserved orientation and provide structural insight into how the acidic C-terminal tail of the primase-helicase contacts the DNA polymerase to facilitate loading of the polymerase onto DNA. A third DNA polymerase binds the ring in an offset manner that may enable polymerase exchange during replication. Alternative polymerase binding modes are also detected by small-angle X-ray scattering with DNA substrates present. Our collective results unveil complex motions within T7 replisome higher-order structures that are underpinned by multivalent protein-protein interactions with functional implications.
- Department of Chemistry & Physics, Western Carolina University, Cullowhee, NC 28723, USA. Electronic address: jamiewallen@email.wcu.edu.
Organizational Affiliation: