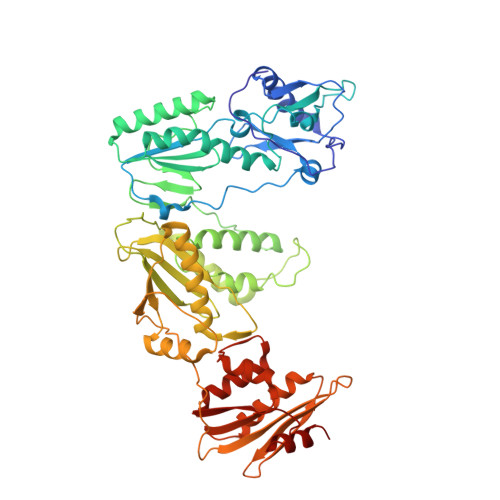
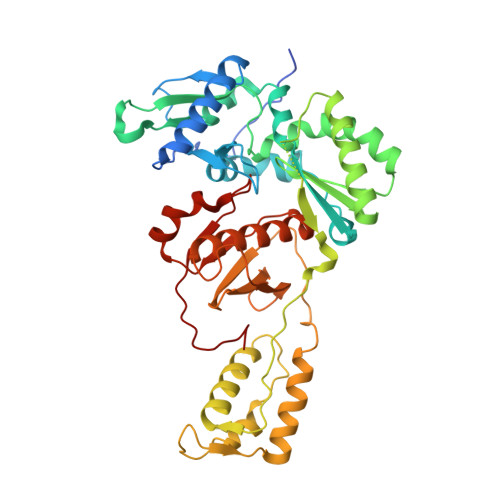
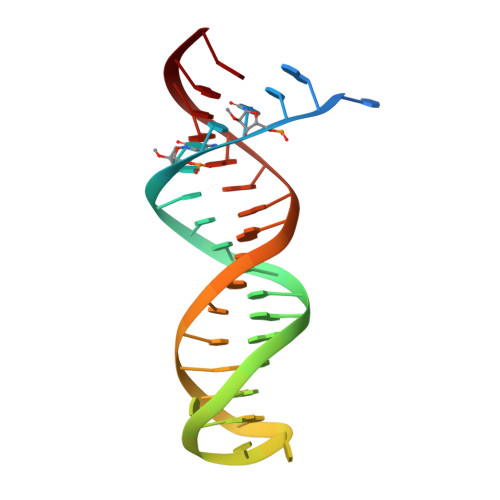
Conformational States of HIV-1 Reverse Transcriptase for Nucleotide Incorporation vs Pyrophosphorolysis-Binding of Foscarnet.
Das, K., Balzarini, J., Miller, M.T., Maguire, A.R., DeStefano, J.J., Arnold, E.(2016) ACS Chem Biol 11: 2158-2164
- PubMed: 27192549
- DOI: https://doi.org/10.1021/acschembio.6b00187
- Primary Citation of Related Structures:
5HP1, 5HRO, 5I3U, 5I42 - PubMed Abstract:
HIV-1 reverse transcriptase (RT) catalytically incorporates individual nucleotides into a viral DNA strand complementing an RNA or DNA template strand; the polymerase active site of RT adopts multiple conformational and structural states while performing this task. The states associated are dNTP binding at the N site, catalytic incorporation of a nucleotide, release of a pyrophosphate, and translocation of the primer 3'-end to the P site. Structural characterization of each of these states may help in understanding the molecular mechanisms of drug activity and resistance and in developing new RT inhibitors. Using a 38-mer DNA template-primer aptamer as the substrate mimic, we crystallized an RT/dsDNA complex that is catalytically active, yet translocation-incompetent in crystals. The ability of RT to perform dNTP binding and incorporation in crystals permitted obtaining a series of structures: (I) RT/DNA (P-site), (II) RT/DNA/AZTTP ternary, (III) RT/AZT-terminated DNA (N-site), and (IV) RT/AZT-terminated DNA (N-site)/foscarnet complexes. The stable N-site complex permitted the binding of foscarnet as a pyrophosphate mimic. The Mg(2+) ions dissociated after catalytic addition of AZTMP in the pretranslocated structure III, whereas ions A and B had re-entered the active site to bind foscarnet in structure IV. The binding of foscarnet involves chelation with the Mg(2+) (B) ion and interactions with K65 and R72. The analysis of interactions of foscarnet and the recently discovered nucleotide-competing RT inhibitor (NcRTI) α-T-CNP in two different conformational states of the enzyme provides insights for developing new classes of polymerase active site RT inhibitors.
- Center for Advanced Biotechnology and Medicine (CABM), Department of Chemistry and Chemical Biology, Rutgers University , Piscataway, New Jersey, United States.
Organizational Affiliation: