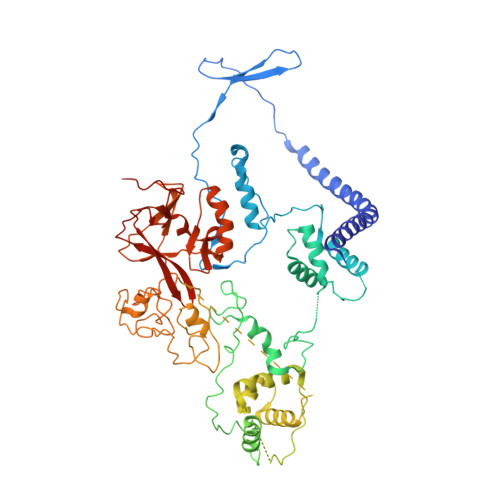
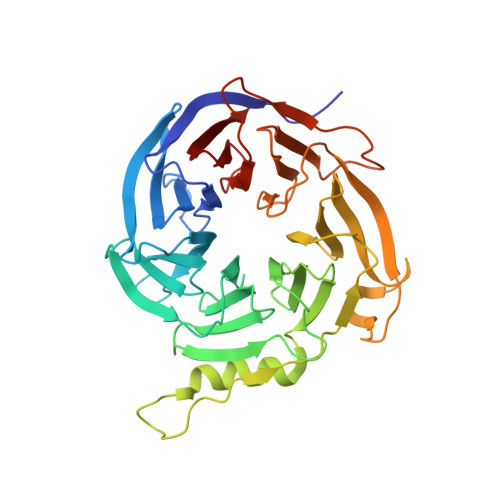
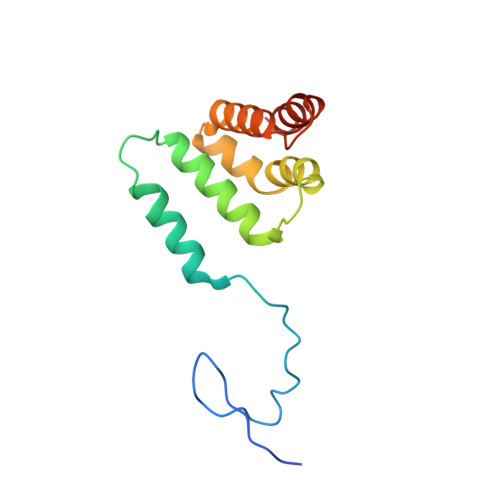
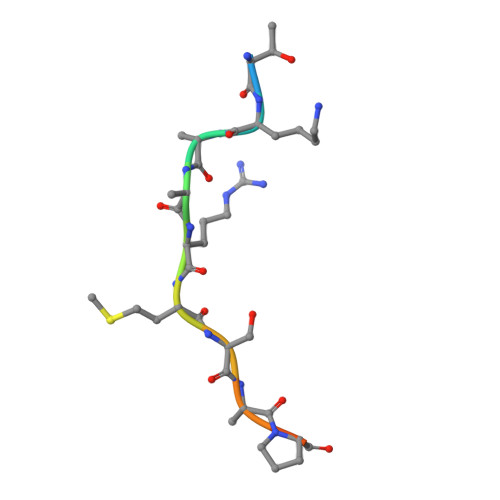
Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2.
Justin, N., Zhang, Y., Tarricone, C., Martin, S.R., Chen, S., Underwood, E., De Marco, V., Haire, L.F., Walker, P.A., Reinberg, D., Wilson, J.R., Gamblin, S.J.(2016) Nat Commun 7: 11316-11316
- PubMed: 27121947
- DOI: https://doi.org/10.1038/ncomms11316
- Primary Citation of Related Structures:
5HYN - PubMed Abstract:
Polycomb repressive complex 2 (PRC2) silences gene expression through trimethylation of K27 of histone H3 (H3K27me3) via its catalytic SET domain. A missense mutation in the substrate of PRC2, histone H3K27M, is associated with certain pediatric brain cancers and is linked to a global decrease of H3K27me3 in the affected cells thought to be mediated by inhibition of PRC2 activity. We present here the crystal structure of human PRC2 in complex with the inhibitory H3K27M peptide bound to the active site of the SET domain, with the methionine residue located in the pocket that normally accommodates the target lysine residue. The structure and binding studies suggest a mechanism for the oncogenic inhibition of H3K27M. The structure also reveals how binding of repressive marks, like H3K27me3, to the EED subunit of the complex leads to enhancement of the catalytic efficiency of the SET domain and thus the propagation of this repressive histone modification.
- The Francis Crick Institute, Mill Hill Laboratory, London NW7 1AA, UK.
Organizational Affiliation: