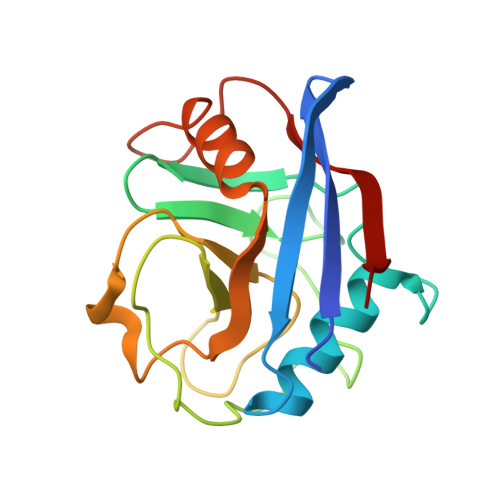
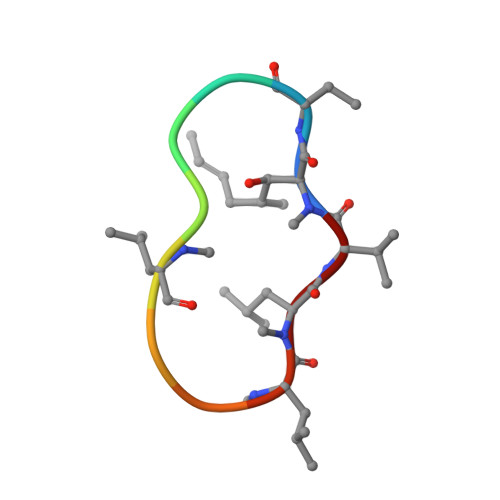
X-ray structure of alisporivir in complex with cyclophilin A at 1.5 angstrom resolution.
Dujardin, M., Bouckaert, J., Rucktooa, P., Hanoulle, X.(2018) Acta Crystallogr F Struct Biol Commun 74: 583-592
- PubMed: 30198892
- DOI: https://doi.org/10.1107/S2053230X18010415
- Primary Citation of Related Structures:
5HSV - PubMed Abstract:
Alisporivir (ALV) is an 11-amino-acid hydrophobic cyclic peptide with N-methyl-D-alanine and N-ethyl-L-valine (NEV) residues at positions 3 and 4, respectively. ALV is a non-immunosuppressive cyclosporin A (CsA) derivative. This inhibitor targets cyclophilins (Cyps), a family of proteins with peptidyl-prolyl cis/trans isomerase enzymatic activity. Cyps act as protein chaperones and are involved in numerous cellular functions. Moreover, Cyps have been shown to be an essential cofactor for the replication of many viruses, including Hepatitis C virus and Human immunodeficiency virus, and have also been shown to be involved in mitochondrial diseases. For these reasons, cyclophilins represent an attractive drug target. The structure of ALV in complex with cyclophilin A (CypA), the most abundant Cyp in humans, has been determined at 1.5 Å resolution. This first structure of the CypA-ALV complex shows that the binding of ALV is highly similar to that of CsA. The high resolution allowed the unambiguous determination of the conformations of residues 3 and 4 in ALV when bound to its target. In particular, the side-chain conformation of NEV4 precludes the interaction of the CypA-ALV complex with calcineurin, a cellular protein phosphatase involved in the immune response, which explains the non-immunosuppressive property of ALV. This study provides detailed molecular insights into the CypA-ALV interaction.
- UGSF-UMR8576, University of Lille, CNRS, F-59000 Lille, France.
Organizational Affiliation: