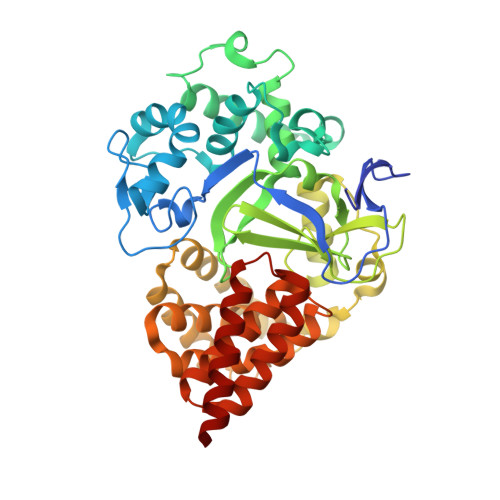
Structure-Based Design of a Novel SMYD3 Inhibitor that Bridges the SAM-and MEKK2-Binding Pockets.
Van Aller, G.S., Graves, A.P., Elkins, P.A., Bonnette, W.G., McDevitt, P.J., Zappacosta, F., Annan, R.S., Dean, T.W., Su, D.S., Carpenter, C.L., Mohammad, H.P., Kruger, R.G.(2016) Structure 24: 774-781
- PubMed: 27066749
- DOI: https://doi.org/10.1016/j.str.2016.03.010
- Primary Citation of Related Structures:
5HI7, 5HQ8 - PubMed Abstract:
SMYD3 is a lysine methyltransferase overexpressed in colorectal, breast, prostate, and hepatocellular tumors, and has been implicated as an oncogene in human malignancies. Methylation of MEKK2 by SMYD3 is important for regulation of the MEK/ERK pathway, suggesting the possibility of selectively targeting SMYD3 in RAS-driven cancers. Structural and kinetic characterization of SMYD3 was undertaken leading to a co-crystal structure of SMYD3 with a MEKK2-peptide substrate bound, and the observation that SMYD3 follows a partially processive mechanism. These insights allowed for the design of GSK2807, a potent and selective, SAM-competitive inhibitor of SMYD3 (Ki = 14 nM). A high-resolution crystal structure reveals that GSK2807 bridges the gap between the SAM-binding pocket and the substrate lysine tunnel of SMYD3. Taken together, our data demonstrate that small-molecule inhibitors of SMYD3 can be designed to prevent methylation of MEKK2 and these could have potential use as anticancer therapeutics.
- Cancer Epigenetics Discovery Performance Unit, GlaxoSmithKline, Collegeville, PA 19426, USA.
Organizational Affiliation: