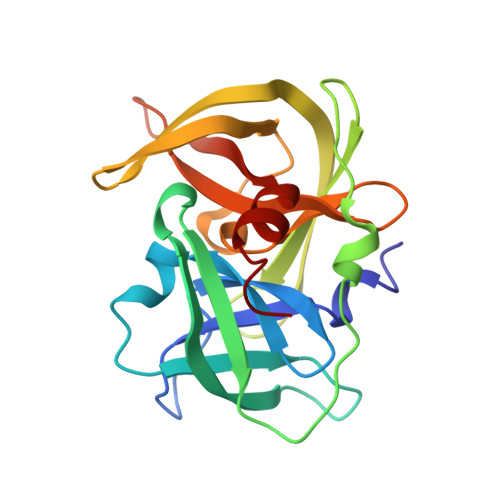
MamO Is a Repurposed Serine Protease that Promotes Magnetite Biomineralization through Direct Transition Metal Binding in Magnetotactic Bacteria.
Hershey, D.M., Ren, X., Melnyk, R.A., Browne, P.J., Ozyamak, E., Jones, S.R., Chang, M.C., Hurley, J.H., Komeili, A.(2016) PLoS Biol 14: e1002402-e1002402
- PubMed: 26981620
- DOI: https://doi.org/10.1371/journal.pbio.1002402
- Primary Citation of Related Structures:
5HM9, 5HMA - PubMed Abstract:
Many living organisms transform inorganic atoms into highly ordered crystalline materials. An elegant example of such biomineralization processes is the production of nano-scale magnetic crystals in magnetotactic bacteria. Previous studies implicated the involvement of two putative serine proteases, MamE and MamO, during the early stages of magnetite formation in Magnetospirillum magneticum AMB-1. Here, using genetic analysis and X-ray crystallography, we show that MamO has a degenerate active site, rendering it incapable of protease activity. Instead, MamO promotes magnetosome formation through two genetically distinct, noncatalytic activities: activation of MamE-dependent proteolysis of biomineralization factors and direct binding to transition metal ions. By solving the structure of the protease domain bound to a metal ion, we identify a surface-exposed di-histidine motif in MamO that contributes to metal binding and show that it is required to initiate biomineralization in vivo. Finally, we find that pseudoproteases are widespread in magnetotactic bacteria and that they have evolved independently in three separate taxa. Our results highlight the versatility of protein scaffolds in accommodating new biochemical activities and provide unprecedented insight into the earliest stages of biomineralization.
- Department of Plant and Microbial Biology, University of California, Berkeley, California, United States of America.
Organizational Affiliation: