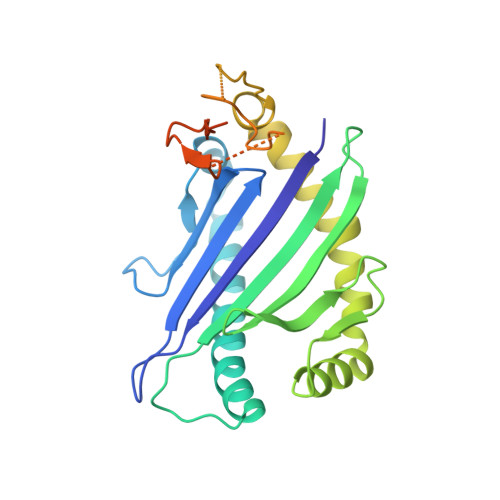
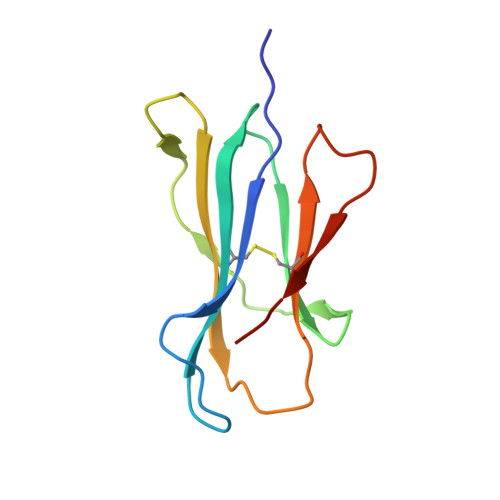
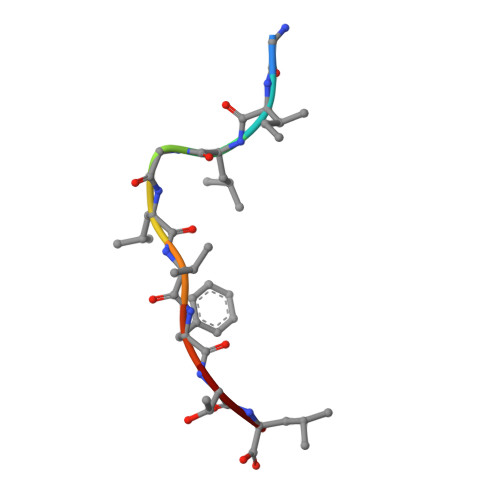
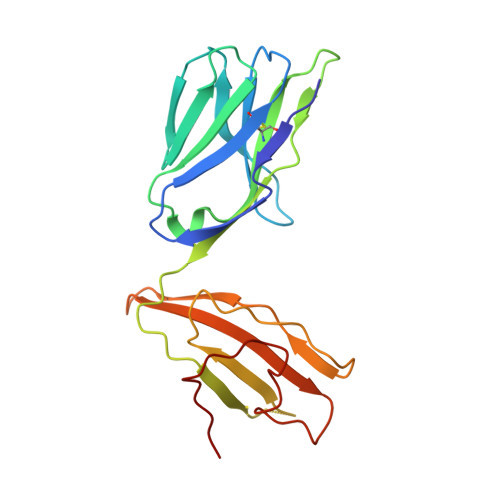
Molecular basis for universal HLA-A*0201-restricted CD8+ T-cell immunity against influenza viruses.
Valkenburg, S.A., Josephs, T.M., Clemens, E.B., Grant, E.J., Nguyen, T.H., Wang, G.C., Price, D.A., Miller, A., Tong, S.Y., Thomas, P.G., Doherty, P.C., Rossjohn, J., Gras, S., Kedzierska, K.(2016) Proc Natl Acad Sci U S A 113: 4440-4445
- PubMed: 27036003
- DOI: https://doi.org/10.1073/pnas.1603106113
- Primary Citation of Related Structures:
5HHM, 5HHN, 5HHO, 5HHP, 5HHQ - PubMed Abstract:
Memory CD8(+)T lymphocytes (CTLs) specific for antigenic peptides derived from internal viral proteins confer broad protection against distinct strains of influenza A virus (IAV). However, immune efficacy can be undermined by the emergence of escape mutants. To determine how T-cell receptor (TCR) composition relates to IAV epitope variability, we used ex vivo peptide-HLA tetramer enrichment and single-cell multiplex analysis to compare TCRs targeted to the largely conserved HLA-A*0201-M158and the hypervariable HLA-B*3501-NP418antigens. The TCRαβs for HLA-B*3501-NP418 (+)CTLs varied among individuals and across IAV strains, indicating that a range of mutated peptides will prime different NP418-specific CTL sets. Conversely, a dominant public TRAV27/TRBV19(+)TCRαβ was selected in HLA-A*0201(+)donors responding to M158 This public TCR cross-recognized naturally occurring M158variants complexed with HLA-A*0201. Ternary structures showed that induced-fit molecular mimicry underpins TRAV27/TRBV19(+)TCR specificity for the WT and mutant M158peptides, suggesting the possibility of universal CTL immunity in HLA-A*0201(+)individuals. Combined with the high population frequency of HLA-A*0201, these data potentially explain the relative conservation of M158 Moreover, our results suggest that vaccination strategies aimed at generating broad protection should incorporate variant peptides to elicit cross-reactive responses against other specificities, especially those that may be relatively infrequent among IAV-primed memory CTLs.
- Department of Microbiology and Immunology, University of Melbourne, at the Peter Doherty Institute for Infection and Immunity, Melbourne, VIC 3000, Australia;
Organizational Affiliation: