Molecular insights into catalysis and processive exonucleolytic mechanisms of prokaryotic RNase J revealing striking parallels with that of eukaryotic Xrn1
Zheng, X., Feng, N., Li, D.F., Li, J., Dong, X.Z.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Ribonuclease J | 470 | Methanolobus psychrophilus R15 | Mutation(s): 1 Gene Names: rnj, Mpsy_0886 EC: 3.1 | 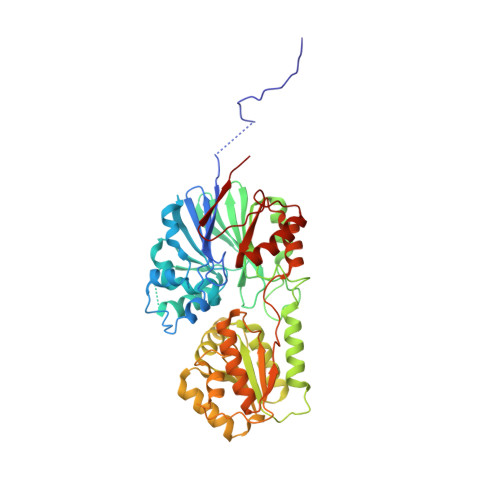 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| RNA (5'-R(P*AP*AP*AP*AP*A)-3') | 5 | synthetic construct | 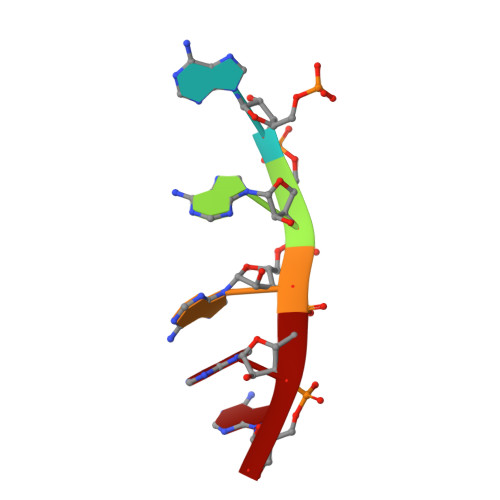 | ||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SO4 Query on SO4 | E [auth A] F [auth A] G [auth A] H [auth A] I [auth B] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 169.68 | α = 90 |
| b = 169.68 | β = 90 |
| c = 167.32 | γ = 120 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| MOSFLM | data reduction |
| SCALA | data scaling |
| PHASER | phasing |